 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_53321.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 142aa MW: 16127.2 Da PI: 9.7824 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 44.8 | 2.6e-14 | 60 | 112 | 3 | 55 |
XXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 3 elkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkke 55
+ +r++r++kNRe+A rsR+RK+a+i+eLe v +Le+e +L e ee +++
MLOC_53321.1 60 ATQRQKRMIKNRESAARSRERKQAYIAELEAQVTQLEEEHAELLREQEEQNEK 112
679*************************************9988776655544 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 6.3E-10 | 57 | 123 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.6E-12 | 60 | 119 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.944 | 60 | 124 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14707 | 1.27E-16 | 62 | 109 | No hit | No description |
| SuperFamily | SSF57959 | 1.48E-11 | 63 | 110 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 5.4E-14 | 63 | 119 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 65 | 80 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 142 aa Download sequence Send to blast |
MTLEDFLARE DDGMVTAVEG NMAVGFPDVG AGVAGGRRRG GGAGGRARKR ALMDPMDRAA 60 TQRQKRMIKN RESAARSRER KQAYIAELEA QVTQLEEEHA ELLREQEEQN EKRLNELKEQ 120 AFQVVVRKKP SQDLRRTNSM EW |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 34 | 41 | GGRRRGGG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the ABA-responsive elements (ABREs) in vitro. Involved in abiotic stress responses and abscisic acid (ABA) signaling (PubMed:20039193). Involved in the signaling pathway that induces growth inhibition in response to D-allose (PubMed:23397192). {ECO:0000269|PubMed:20039193, ECO:0000269|PubMed:23397192}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00648 | PBM | Transfer from LOC_Os05g36160 | Download |
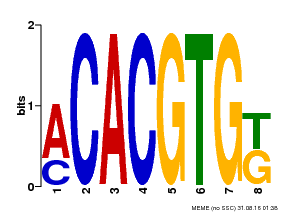
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_53321.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by anoxia, drought, salt stress, oxidative stress, cold and abscisic acid (ABA) (PubMed:20039193). Induced by D-allose (PubMed:23397192). {ECO:0000269|PubMed:20039193, ECO:0000269|PubMed:23397192}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK367259 | 0.0 | AK367259.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2053K07. | |||
| GenBank | AK368116 | 0.0 | AK368116.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2068G17. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020178558.1 | 1e-75 | G-box-binding factor 4-like | ||||
| Swissprot | Q0JHF1 | 1e-55 | BZP12_ORYSJ; bZIP transcription factor 12 | ||||
| TrEMBL | F2DTP1 | 3e-96 | F2DTP1_HORVV; Predicted protein | ||||
| STRING | MLOC_53321.1 | 2e-97 | (Hordeum vulgare) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2665 | 37 | 86 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G44080.1 | 2e-17 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



