 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_54829.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 453aa MW: 49355.7 Da PI: 5.3235 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.6 | 1e-31 | 274 | 328 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k rlrWt eLHerFveav++L G++kAtPk +l+lmkv+gLt++hvkSHLQkYR
MLOC_54829.1 274 KSRLRWTLELHERFVEAVNKLEGPDKATPKGVLKLMKVDGLTIYHVKSHLQKYRH 328
68****************************************************6 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.269 | 271 | 331 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.3E-30 | 272 | 329 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 7.17E-17 | 273 | 328 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.1E-24 | 274 | 328 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.7E-8 | 276 | 327 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 2.5E-21 | 363 | 408 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 453 aa Download sequence Send to blast |
MSSHGVVAVK PIAAPEKTAH SYACGSTQSS VHKLLDAKLD HLGLLDDNLS STSQSSDIKT 60 ELIQSSSLGR SSLPFNLQRR SPEPDPESPL SHVSHPNFSE PMASNSSTFC TSLFSSSSTN 120 SAPCRQMGAL PFLPHPPKYE QQVLPGQSSN SSLQLSGDTG NVHDETEQPD DIKDFLNLSA 180 GDASDASFHG ENQAFAFAED EQMEFQFLSE QLGIAITDNE ESPQLDDIYD TPPPQMLSLP 240 VSSCSNQSLQ NPGSPGKLPL SSSRSSSGSA AANKSRLRWT LELHERFVEA VNKLEGPDKA 300 TPKGVLKLMK VDGLTIYHVK SHLQKYRHAK FIPEIKEEKK ASLDVKKVQP GSSGSDPFKN 360 KNLAEALRMQ LEVQKQLHEQ LEVQRQLQLR IEEHAKYLQR ILEEQQKAIA SSGSSLSLKT 420 PTEPSESTSK DGAEPEEATT SSPQPSKNSE TES |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 3e-24 | 274 | 331 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 3e-24 | 274 | 331 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 3e-24 | 274 | 331 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 3e-24 | 274 | 331 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 3e-24 | 274 | 331 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 3e-24 | 274 | 331 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 3e-24 | 274 | 331 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 3e-24 | 274 | 331 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in phosphate starvation signaling (PubMed:26082401). Binds to P1BS, an imperfect palindromic sequence 5'-GNATATNC-3', to promote the expression of inorganic phosphate (Pi) starvation-responsive genes (PubMed:26082401). Functionally redundant with PHR1 and PHR2 in regulating Pi starvation response and Pi homeostasis (PubMed:26082401). {ECO:0000269|PubMed:26082401}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00354 | DAP | Transfer from AT3G13040 | Download |
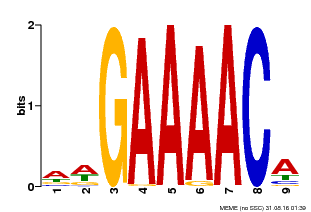
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_54829.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated under Pi starvation conditions. {ECO:0000269|PubMed:26082401}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK250795 | 0.0 | AK250795.1 Hordeum vulgare subsp. vulgare cDNA clone: FLbaf91j11, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020163203.1 | 0.0 | protein PHOSPHATE STARVATION RESPONSE 3-like | ||||
| Refseq | XP_020163204.1 | 0.0 | protein PHOSPHATE STARVATION RESPONSE 3-like | ||||
| Swissprot | Q6YXZ4 | 1e-145 | PHR3_ORYSJ; Protein PHOSPHATE STARVATION RESPONSE 3 | ||||
| TrEMBL | M0WUM8 | 0.0 | M0WUM8_HORVV; Uncharacterized protein | ||||
| STRING | MLOC_54829.2 | 0.0 | (Hordeum vulgare) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4526 | 37 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G13040.2 | 4e-61 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



