 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_57220.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 235aa MW: 26003.2 Da PI: 9.9129 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 58.4 | 1.6e-18 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rgrWT+eEd++l+++++q+G g+W++ ++ g+ R++k+c++rw +yl
MLOC_57220.2 14 RGRWTAEEDDILANYIAQHGEGSWRSLPKNAGLLRCGKSCRLRWINYL 61
8*******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 56.9 | 4.6e-18 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+ ++eEd+l+v++++ lG++ W++Ia++++ gRt++++k++w+++l
MLOC_57220.2 67 RGNISKEEDDLIVKLHATLGNR-WSLIASHLP-GRTDNEIKNYWNSHL 112
7899******************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 7.4E-22 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.64 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.76E-29 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.1E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.3E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.02E-11 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 25.884 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-25 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.8E-17 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.6E-16 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.85E-13 | 71 | 112 | No hit | No description |
| Pfam | PF06640 | 4.4E-10 | 130 | 191 | IPR010588 | Myb-related protein P, C-terminal |
| Pfam | PF06640 | 1.8E-8 | 192 | 235 | IPR010588 | Myb-related protein P, C-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009813 | Biological Process | flavonoid biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 235 aa Download sequence Send to blast |
MGRAPCCEKV GLKRGRWTAE EDDILANYIA QHGEGSWRSL PKNAGLLRCG KSCRLRWINY 60 LRDGVRRGNI SKEEDDLIVK LHATLGNRWS LIASHLPGRT DNEIKNYWNS HLSRQIHTFR 120 RIYTTVRDTT ITVDIHKLSA AGKRRGGRTP GQSPKSSTKK KQPVPEPVTN AKGTSSTASS 180 PPQSDEARSA VGFASNLWDQ PAQNGLLQPA EPQVATGSES DELESFVSWL LSDAC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 1e-23 | 12 | 118 | 5 | 110 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor postulated to regulate the biosynthetic pathway of a flavonoid-derived pigment in certain floral tissues. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00627 | PBM | Transfer from PK06182.1 | Download |
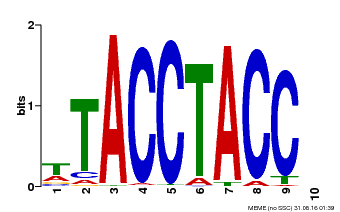
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_57220.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK356334 | 0.0 | AK356334.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1033F20. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020199233.1 | 1e-117 | myb-related protein P-like isoform X1 | ||||
| Swissprot | P27898 | 3e-86 | MYBP_MAIZE; Myb-related protein P | ||||
| TrEMBL | M0X5L3 | 1e-173 | M0X5L3_HORVV; Uncharacterized protein | ||||
| STRING | MLOC_57220.1 | 1e-138 | (Hordeum vulgare) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47460.1 | 5e-71 | myb domain protein 12 | ||||



