 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_58019.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 274aa MW: 29335.7 Da PI: 8.1111 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 105.9 | 2.1e-33 | 86 | 144 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct++gC+v+k+ver+++d k v++tYeg+Hnhe
MLOC_58019.1 86 LDDGYRWRKYGQKVVKGNPNPRSYYKCTHQGCSVRKHVERASHDLKSVITTYEGKHNHE 144
59********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 7.4E-38 | 71 | 146 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.57E-29 | 78 | 146 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 37.883 | 81 | 146 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.3E-38 | 86 | 145 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 7.3E-26 | 87 | 144 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0030010 | Biological Process | establishment of cell polarity | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 274 aa Download sequence Send to blast |
MESQDAADVS STLSNEIDRA TQGTISLDCD GGEDETESKR RKLDALAAVT LPTATTTSSI 60 DMVAAASRAV REPRVVVQTT SEVDILDDGY RWRKYGQKVV KGNPNPRSYY KCTHQGCSVR 120 KHVERASHDL KSVITTYEGK HNHEVPAARN SGNGGSGSGS AQPSAPQANI SHRRQEQAQG 180 SYPQFGGATP FGSFGLPPRG HLGAAGNFHF GMGPPGMSMP PMPAARHPPS MMQGYPGLMM 240 QEGQMKPEPD QQSGFASASA YQQMMGRPPF GPQM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 3e-38 | 77 | 147 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 3e-38 | 77 | 147 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Regulates WOX8 and WOX9 expression and basal cell division patterns during early embryogenesis. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Required to repolarize the zygote from a transient symmetric state. {ECO:0000269|PubMed:21316593}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00071 | PBM | Transfer from AT5G56270 | Download |
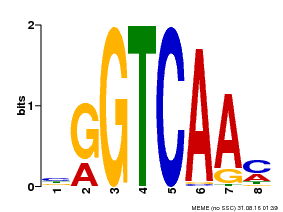
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_58019.1 |
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK362158 | 0.0 | AK362158.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2002O24. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020200291.1 | 0.0 | probable WRKY transcription factor 2 | ||||
| Swissprot | Q9FG77 | 1e-62 | WRKY2_ARATH; Probable WRKY transcription factor 2 | ||||
| TrEMBL | A0A453L317 | 0.0 | A0A453L317_AEGTS; Uncharacterized protein | ||||
| STRING | MLOC_58019.2 | 1e-178 | (Hordeum vulgare) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56270.1 | 4e-51 | WRKY DNA-binding protein 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



