 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_6041.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 427aa MW: 46251.5 Da PI: 5.3954 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.8 | 1.8e-16 | 42 | 89 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+ Ed llv+ v+q+G g+W+++ r g+ R++k+c++rw ++l
MLOC_6041.1 42 KGPWTAAEDALLVNHVRQHGEGNWNAVQRITGLLRCGKSCRLRWTNHL 89
79******************************99***********996 PP
| |||||||
| 2 | Myb_DNA-binding | 51.3 | 2.7e-16 | 95 | 138 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g+++++E++l++++++qlG++ W++ a +++ gRt++++k++w++
MLOC_6041.1 95 KGAFSPDEELLIAQLHAQLGNK-WARMATHLP-GRTDNEIKNYWNT 138
799*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.186 | 37 | 89 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.6E-14 | 41 | 91 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.35E-28 | 41 | 136 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 4.7E-14 | 42 | 89 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.1E-21 | 43 | 96 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.84E-11 | 44 | 89 | No hit | No description |
| PROSITE profile | PS51294 | 23.627 | 90 | 144 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.5E-14 | 94 | 142 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.0E-15 | 95 | 138 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-23 | 97 | 143 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.08E-10 | 97 | 138 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0090406 | Cellular Component | pollen tube | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 427 aa Download sequence Send to blast |
MDAASLEEEK MAAAAAAADN LDEEAEQGGE GEEEAMPVVL KKGPWTAAED ALLVNHVRQH 60 GEGNWNAVQR ITGLLRCGKS CRLRWTNHLR PNLKKGAFSP DEELLIAQLH AQLGNKWARM 120 ATHLPGRTDN EIKNYWNTRA KRRQRAGLPM YPPEVQFQLA ITKRCRYDDF TPPQQSAGGN 180 VLDATDARYT SARPPPLDLA GQLAMANRPV QFLAQTPFSA PSSPWVKPSF ARNARYFQFP 240 HSSPVSPTTP VHPVTPELSL GHGQHGGERS RFTPLSPSPG AKAELPSGQL LPASPAAATP 300 TSGEDQQNLE AMLQELHDAI KIEPPKHVSD HAEQDGGSGG NKPEGELKDD DIGTLLDMII 360 PATFPMTEPA PPGVAPDNSG GSISQNGSND HDHHNVDLSV DHIPIISSEQ DWVLDGACQW 420 NNMSGIC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 6e-27 | 40 | 143 | 25 | 127 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00287 | DAP | Transfer from AT2G32460 | Download |
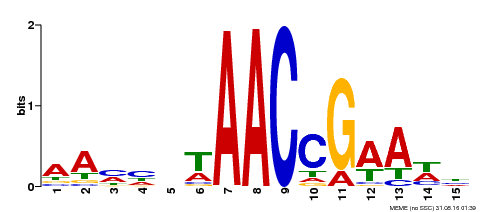
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_6041.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT042537 | 1e-122 | BT042537.1 Zea mays full-length cDNA clone ZM_BFb0325G13 mRNA, complete cds. | |||
| GenBank | KJ727532 | 1e-122 | KJ727532.1 Zea mays clone pUT5392 MYB transcription factor (MYB129) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020182637.1 | 0.0 | uncharacterized protein LOC109768314 | ||||
| TrEMBL | M0XIM7 | 0.0 | M0XIM7_HORVV; Uncharacterized protein | ||||
| STRING | MLOC_6041.1 | 0.0 | (Hordeum vulgare) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7420 | 37 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G32460.1 | 7e-66 | myb domain protein 101 | ||||



