 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_63066.11 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 153aa MW: 17252.2 Da PI: 9.5271 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 68.4 | 1.1e-21 | 56 | 118 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e+elkr++rkq+NR +ArrsR+RK+ae+eeL++++++L++eN +Lk+e+++++ke+++l s++
MLOC_63066.11 56 ERELKRQKRKQSNRDSARRSRLRKQAECEELAQRAEVLKQENASLKDEVSRIRKEYDELLSKN 118
89***********************************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 2.4E-18 | 50 | 115 | No hit | No description |
| SMART | SM00338 | 5.2E-21 | 56 | 120 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 9.7E-21 | 56 | 118 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 13.082 | 58 | 121 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.63E-11 | 59 | 115 | No hit | No description |
| CDD | cd14702 | 4.45E-19 | 61 | 112 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 63 | 78 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 153 aa Download sequence Send to blast |
MQMPSSGPVP GPTTNLKIGM DYWANTASSS PALHGKVTPT AIPGDLAPTE PWMQDERELK 60 RQKRKQSNRD SARRSRLRKQ AECEELAQRA EVLKQENASL KDEVSRIRKE YDELLSKNSS 120 LKDNIGDKQH KTDEAGLHNK LQRSGDDNQK DTN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 72 | 78 | RRSRLRK |
| 2 | 72 | 79 | RRSRLRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in responses to fungal pathogen infection and abiotic stresses. {ECO:0000269|Ref.1}. | |||||
| UniProt | Probable transcription factor that may be involved in responses to fungal pathogen infection and abiotic stresses. {ECO:0000269|Ref.2}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00291 | DAP | Transfer from AT2G35530 | Download |
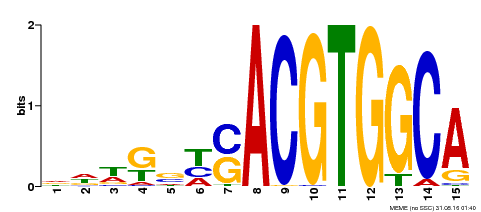
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_63066.11 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced during incompatible interaction with the fungal pathogen Puccinia striiformis (Ref.1). Induced by abscisic acid (ABA), ethylene, cold stress, salt stress and wounding (Ref.1). {ECO:0000269|Ref.1}. | |||||
| UniProt | INDUCTION: Induced during incompatible interaction with the fungal pathogen Puccinia striiformis (Ref.2). Induced by abscisic acid (ABA), ethylene, cold stress, salt stress and wounding (Ref.2). {ECO:0000269|Ref.2}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK366562 | 0.0 | AK366562.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2044E07. | |||
| GenBank | AK373997 | 0.0 | AK373997.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3050F17. | |||
| GenBank | AY150677 | 0.0 | AY150677.1 Hordeum vulgare subsp. vulgare bZIP transcription factor ZIP1 (ZIP1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020189928.1 | 1e-102 | bZIP transcription factor 16-like | ||||
| Swissprot | A0A3B6MPP5 | 1e-103 | BZP1D_WHEAT; bZIP transcription factor 1-D | ||||
| Swissprot | B6E107 | 1e-103 | BZP1B_WHEAT; bZIP transcription factor 1-B | ||||
| TrEMBL | A0A287QSB6 | 1e-108 | A0A287QSB6_HORVV; Uncharacterized protein | ||||
| TrEMBL | M0XU65 | 1e-109 | M0XU65_HORVV; Uncharacterized protein | ||||
| TrEMBL | M0XU72 | 1e-108 | M0XU72_HORVV; Uncharacterized protein | ||||
| STRING | TRIUR3_34165-P1 | 1e-102 | (Triticum urartu) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G35530.1 | 5e-48 | basic region/leucine zipper transcription factor 16 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



