 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_64817.3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 325aa MW: 34450.9 Da PI: 9.8124 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 55.9 | 9.6e-18 | 19 | 64 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+g+W++eEd+ l ++v ++G ++W++I++ ++ gR++k+c++rw +
MLOC_64817.3 19 KGPWSPEEDDALQRLVGRHGARNWSLISKSIP-GRSGKSCRLRWCNQ 64
79******************************.***********985 PP
| |||||||
| 2 | Myb_DNA-binding | 51.9 | 1.8e-16 | 73 | 115 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
++T++Ed+ +++a++++G++ W+tIar + gRt++ +k++w++
MLOC_64817.3 73 PFTPDEDDTILRAHARFGNK-WATIARLLS-GRTDNAIKNHWNST 115
89******************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 25.716 | 14 | 69 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.88E-31 | 16 | 112 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.0E-15 | 18 | 67 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.4E-17 | 19 | 64 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.4E-25 | 20 | 72 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.62E-15 | 21 | 63 | No hit | No description |
| SMART | SM00717 | 7.2E-14 | 70 | 118 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 19.747 | 71 | 120 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.0E-23 | 73 | 119 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.80E-11 | 73 | 116 | No hit | No description |
| Pfam | PF00249 | 5.3E-14 | 73 | 115 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 325 aa Download sequence Send to blast |
MAACRGGGGG GGKDVDRIKG PWSPEEDDAL QRLVGRHGAR NWSLISKSIP GRSGKSCRLR 60 WCNQLSPQVE HRPFTPDEDD TILRAHARFG NKWATIARLL SGRTDNAIKN HWNSTLKRKY 120 SSSSSASASA SASPGDDAVD DDRRPLKRTS SDGYPGLCFS PGSPSGSDLS DSSHQSLPSV 180 MPSAASHQPH VYRPVARAGG VVVLPTATPQ PSPPSPPPPP PPPPPATSLS LSLSLPGLDA 240 PEPAPVAQPM PMPMPPPPVP APVQPAAAPQ MPPPSMPFHL QPPPPPQPRA AAPFNGEFLS 300 MMQEMIRIEV RNYMSGFHPR SPTSA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 1e-38 | 18 | 119 | 3 | 104 | MYB PROTO-ONCOGENE PROTEIN |
| 1mse_C | 1e-38 | 18 | 119 | 3 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 1e-38 | 18 | 119 | 3 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 4 | 12 | RGGGGGGGK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00476 | DAP | Transfer from AT4G37260 | Download |
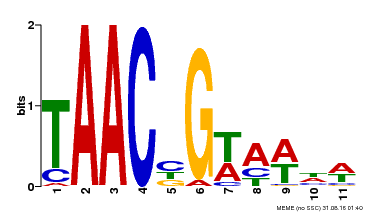
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_64817.3 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK355241 | 0.0 | AK355241.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1018A13. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020146152.1 | 1e-161 | transcription factor MYB44-like | ||||
| TrEMBL | F2CUD9 | 0.0 | F2CUD9_HORVV; Predicted protein | ||||
| STRING | Traes_5DL_92CBBE968.1 | 1e-160 | (Triticum aestivum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G23290.1 | 1e-68 | myb domain protein 70 | ||||



