 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_65876.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 315aa MW: 34384.9 Da PI: 9.6314 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 77.9 | 1.3e-24 | 13 | 66 | 3 | 56 |
G2-like 3 rlrWtpeLHerFveaveqLGGsekAtPktilelmkvk.gLtlehvkSHLQkYRla 56
+++WtpeLH+rFv+aveqL G +kA+P++ilelm+ + Lt+++++SHLQkYR++
MLOC_65876.1 13 QVDWTPELHRRFVQAVEQL-GLDKAVPSRILELMGNEyRLTRHNIASHLQKYRSH 66
689****************.**************88747**************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.412 | 8 | 68 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.35E-16 | 12 | 69 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.8E-24 | 12 | 69 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 8.7E-24 | 13 | 66 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 7.9E-9 | 15 | 64 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007165 | Biological Process | signal transduction | ||||
| GO:0009658 | Biological Process | chloroplast organization | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0010380 | Biological Process | regulation of chlorophyll biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 315 aa Download sequence Send to blast |
MAVGSADEMM ESQVDWTPEL HRRFVQAVEQ LGLDKAVPSR ILELMGNEYR LTRHNIASHL 60 QKYRSHRKHL MAREAEAASW THKRQMYAAA GGPRKDAPAG GGPWVVPTVG FPPPGTMPHP 120 HAAMAHHPGQ PPPFCRPLHV WGHPTGVDAP LPLPLSPPST MLPVWPRHLA PPPAWAHQPP 180 VDPVYWHQQY NAARKWGPQA VTQCVPPPMP PAAMMQRFAA PPMPGMMPHP MYRPIPPPPS 240 PVPQNNKVAG LQLQLDAHPS KESIDAAIGD VLVKPWLPLP LGLKPPSLDS VMSELHKQGI 300 PKVPPAATTN CDGAA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional activator that promotes chloroplast development. Acts as an activator of nuclear photosynthetic genes involved in chlorophyll biosynthesis, light harvesting, and electron transport (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00022 | PBM | Transfer from AT2G20570 | Download |
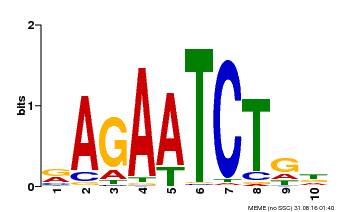
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_65876.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By light. {ECO:0000269|PubMed:11340194}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353571 | 0.0 | AK353571.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1001B16. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020187877.1 | 1e-177 | probable transcription factor GLK2 | ||||
| Swissprot | Q5NAN5 | 1e-88 | GLK2_ORYSJ; Probable transcription factor GLK2 | ||||
| TrEMBL | M0Y5G8 | 0.0 | M0Y5G8_HORVV; Uncharacterized protein | ||||
| STRING | MLOC_65876.1 | 0.0 | (Hordeum vulgare) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2415 | 38 | 86 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G20570.1 | 2e-51 | GBF's pro-rich region-interacting factor 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



