 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_68189.8 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 164aa MW: 18330 Da PI: 10.5241 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 44 | 5e-14 | 35 | 79 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT++E++++++a +++ + Wk+I + +g +t q++s+ qky
MLOC_68189.8 35 RESWTEQEHDKFLEALQLFDRD-WKKIEAFVG-SKTVIQIRSHAQKY 79
789*****************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.84E-17 | 29 | 85 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 15.408 | 30 | 84 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.3E-7 | 33 | 85 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.4E-19 | 33 | 82 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.3E-11 | 34 | 82 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 8.6E-12 | 35 | 79 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.23E-9 | 37 | 80 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 164 aa Download sequence Send to blast |
MVSTNPPPPP ALSDAAAGDD ASKKVRKPYT ITKSRESWTE QEHDKFLEAL QLFDRDWKKI 60 EAFVGSKTVI QIRSHAQKYF LKVQKNGTSE HVPPPRPKRK AAHPYPQKAS KNEPGYALKT 120 DPSAMLRNSG MNVAVSSWTH NSIPPVVASS FMKGLYSKVP LLYK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator of evening element (EE)-containing clock-controlled genes. Forms a negative feedback loop with APRR5. Regulates the pattern of histone H3 acetylation of the TOC1 promoter. {ECO:0000269|PubMed:21205033, ECO:0000269|PubMed:21474993, ECO:0000269|PubMed:21483796, ECO:0000269|PubMed:23638299}. | |||||
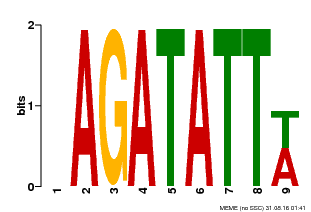
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00625 | PBM | Transfer from PK02532.1 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_68189.8 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation. Peak of expression in the afternoon. Down-regulated by cold. {ECO:0000269|PubMed:21205033, ECO:0000269|PubMed:22902701, ECO:0000269|PubMed:23638299}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK354834 | 0.0 | AK354834.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1011N06. | |||
| GenBank | AK358510 | 0.0 | AK358510.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1077K09. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020182045.1 | 1e-105 | protein REVEILLE 6-like isoform X2 | ||||
| Refseq | XP_020182047.1 | 1e-106 | protein REVEILLE 6-like isoform X4 | ||||
| Swissprot | Q8RWU3 | 7e-63 | RVE8_ARATH; Protein REVEILLE 8 | ||||
| TrEMBL | A0A287UH06 | 1e-112 | A0A287UH06_HORVV; Predicted protein | ||||
| STRING | MLOC_68189.3 | 1e-108 | (Hordeum vulgare) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G52660.2 | 2e-48 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



