 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_69710.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 250aa MW: 27649.7 Da PI: 4.6892 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 61 | 1.8e-19 | 27 | 79 | 4 | 56 |
-SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 4 RttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+ +f++eq++ Le++F+ + ++ +++ +LA++lgL+ rqV +WFqN+Ra++k
MLOC_69710.1 27 KKRFSEEQIKSLESMFATQTKLEPRQKLQLARELGLQPRQVAIWFQNKRARWK 79
456*************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 122.8 | 1.6e-39 | 26 | 117 | 2 | 93 |
HD-ZIP_I/II 2 kkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelke 93
+k+r+s+eq+k+LE++F +++kLep++K +lareLglqprqva+WFqn+RAR+k+kqlE++y+aL++ ydal ++ ++L+k++++L ++l++
MLOC_69710.1 26 RKKRFSEEQIKSLESMFATQTKLEPRQKLQLARELGLQPRQVAIWFQNKRARWKSKQLERQYAALRDDYDALLSSYDQLKKDKQALVNQLEK 117
8***************************************************************************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 4.8E-19 | 22 | 81 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 3.6E-16 | 24 | 85 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 2.05E-18 | 24 | 83 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 16.763 | 26 | 81 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.91E-17 | 26 | 82 | No hit | No description |
| Pfam | PF00046 | 8.0E-17 | 27 | 79 | IPR001356 | Homeobox domain |
| PRINTS | PR00031 | 8.3E-6 | 52 | 61 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 56 | 79 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 8.3E-6 | 61 | 77 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 4.0E-13 | 81 | 122 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 250 aa Download sequence Send to blast |
MEQGEEDGDW MMEPASGKKG GVMIDRKKRF SEEQIKSLES MFATQTKLEP RQKLQLAREL 60 GLQPRQVAIW FQNKRARWKS KQLERQYAAL RDDYDALLSS YDQLKKDKQA LVNQLEKLAE 120 MLREPGGAKC GDNAGAADRD DVRLAVAGMS MKDEFADAAG ASKLYSASAE GCGGSGKLSL 180 FGEDDDDAGL FLRPSLQLPT AHDGGFTASG PAEYQQQSPS SFPFHSSWPS SATEQTCSSS 240 QWWEFESPSE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. {ECO:0000269|PubMed:10732669}. | |||||
| UniProt | Probable transcription factor that binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. {ECO:0000269|PubMed:10732669}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00650 | PBM | Transfer from LOC_Os09g35910 | Download |
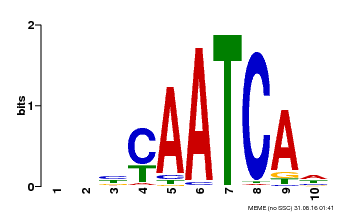
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_69710.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK361212 | 0.0 | AK361212.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, partial cds, clone: NIASHv1135C21. | |||
| GenBank | AK376953 | 0.0 | AK376953.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, partial cds, clone: NIASHv3144J23. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020148448.1 | 1e-165 | homeobox-leucine zipper protein HOX6-like | ||||
| Swissprot | Q651Z5 | 1e-93 | HOX6_ORYSJ; Homeobox-leucine zipper protein HOX6 | ||||
| Swissprot | Q9XH35 | 1e-93 | HOX6_ORYSI; Homeobox-leucine zipper protein HOX6 | ||||
| TrEMBL | M0YIP2 | 0.0 | M0YIP2_HORVV; Uncharacterized protein | ||||
| STRING | MLOC_69710.1 | 0.0 | (Hordeum vulgare) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7873 | 34 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46680.1 | 9e-43 | homeobox 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



