 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_74469.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | SRS | ||||||||
| Protein Properties | Length: 136aa MW: 13853.2 Da PI: 4.5158 | ||||||||
| Description | SRS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF702 | 77.1 | 5e-24 | 5 | 65 | 94 | 154 |
DUF702 94 aeskkeletsslPeevsseavfrcvrvssvddgeeelaYqtavsigGhvfkGiLydqGlee 154
++s+ ++P+e+s eavfrcvr++ vd+ ++e+aYqtavsigGh+fkGiL+d+G++e
MLOC_74469.2 5 TTSSAGEGDGRFPPELSLEAVFRCVRIGPVDEPDAEFAYQTAVSIGGHTFKGILRDHGPAE 65
44555566677***********************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05142 | 8.1E-21 | 9 | 64 | IPR007818 | Protein of unknown function DUF702 |
| TIGRFAMs | TIGR01624 | 3.4E-19 | 15 | 63 | IPR006511 | Lateral Root Primordium type 1, C-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010252 | Biological Process | auxin homeostasis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048479 | Biological Process | style development | ||||
| GO:0048480 | Biological Process | stigma development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 136 aa Download sequence Send to blast |
MVDATTSSAG EGDGRFPPEL SLEAVFRCVR IGPVDEPDAE FAYQTAVSIG GHTFKGILRD 60 HGPAEEAAGQ LPPSSAEYHQ LTGAAREGSS PAGSSEAAGG HGATVATSAA VLMDPYPTPI 120 GAFAAGTQFF PHNPRT |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00613 | PBM | Transfer from AT3G51060 | Download |
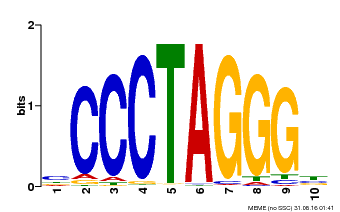
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_74469.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB678347 | 0.0 | AB678347.1 Hordeum vulgare subsp. vulgare Lks2 gene for putative short internodesfamily transcription factor, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020192925.1 | 2e-88 | protein SHI RELATED SEQUENCE 1-like | ||||
| Refseq | XP_020192926.1 | 2e-88 | protein SHI RELATED SEQUENCE 1-like | ||||
| TrEMBL | A0A287XEN8 | 4e-92 | A0A287XEN8_HORVV; Putative short internodesfamily transcription factor | ||||
| STRING | MLOC_74469.1 | 3e-89 | (Hordeum vulgare) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G66350.1 | 8e-24 | Lateral root primordium (LRP) protein-related | ||||



