 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_9943.3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 145aa MW: 16283.2 Da PI: 7.6208 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43.1 | 9.9e-14 | 96 | 144 | 3 | 48 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHT...TTS-HHHHHHHHHHHT CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmg...kgRtlkqcksrwqkyl 48
rW+ E+e l+d+v+q+G+g+Wk I ++ gRt+ ++k++w++ +
MLOC_9943.3 96 RWSSVEEEALKDGVEQFGSGNWKDILSHNAdvfIGRTPVDLKDKWRNMM 144
9********************************9************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.904 | 89 | 145 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.3E-15 | 89 | 145 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 5.6E-16 | 90 | 144 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.0E-5 | 93 | 145 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 1.82E-20 | 95 | 145 | No hit | No description |
| Pfam | PF00249 | 1.3E-10 | 96 | 143 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009901 | Biological Process | anther dehiscence | ||||
| GO:0010152 | Biological Process | pollen maturation | ||||
| GO:0043067 | Biological Process | regulation of programmed cell death | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 145 aa Download sequence Send to blast |
MDNAVNINAE VNNQAANCSI AGSSAVNDQG ETLRKGMPSS LMDWNPTAQS LLWEDSIDSD 60 GSRSQLHRPH LPSPRRIPVS PLQVAENKSM RRRARRWSSV EEEALKDGVE QFGSGNWKDI 120 LSHNADVFIG RTPVDLKDKW RNMMR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00052 | PBM | Transfer from AT1G15720 | Download |
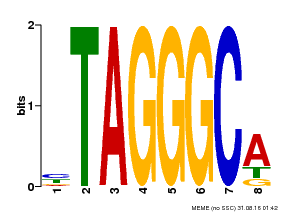
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_9943.3 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK375507 | 0.0 | AK375507.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3096E16. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020185046.1 | 1e-77 | uncharacterized protein LOC109770747 | ||||
| TrEMBL | F2EH80 | 1e-100 | F2EH80_HORVV; Predicted protein | ||||
| STRING | MLOC_9943.1 | 1e-102 | (Hordeum vulgare) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G15720.1 | 3e-16 | TRF-like 5 | ||||



