 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.01G117100.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 240aa MW: 27415.6 Da PI: 10.7739 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 55.6 | 1.2e-17 | 9 | 56 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd ll ++v ++G g W+ ++ + g++R++k+c++rw++yl
Manes.01G117100.1.p 9 KGAWTPEEDNLLRKCVDKYGEGKWHQVPLKAGLNRCRKSCRLRWLNYL 56
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 46.7 | 7.1e-15 | 62 | 107 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g + ++E +l+++++k+lG++ W++Ia +++ gRt++++k++w+++l
Manes.01G117100.1.p 62 KGEFEADEVDLIIRLHKLLGNR-WALIAGRIP-GRTANDVKNYWNTHL 107
688999****************.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 26.379 | 4 | 60 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.44E-29 | 6 | 103 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-22 | 7 | 59 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.4E-15 | 8 | 58 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-16 | 9 | 56 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.92E-11 | 11 | 56 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.9E-23 | 60 | 110 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.3E-15 | 61 | 109 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 19.833 | 61 | 111 | IPR017930 | Myb domain |
| Pfam | PF00249 | 9.8E-14 | 62 | 107 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.40E-10 | 64 | 107 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0031540 | Biological Process | regulation of anthocyanin biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 240 aa Download sequence Send to blast |
MEGLFGVRKG AWTPEEDNLL RKCVDKYGEG KWHQVPLKAG LNRCRKSCRL RWLNYLKPNI 60 KKGEFEADEV DLIIRLHKLL GNRWALIAGR IPGRTANDVK NYWNTHLRKK EASARINAKK 120 HSTPTMTKAS IIKPRPFNLP KKVFWSSNGK TPVTLTTHVP VIDNNNTLCK PCMPSPPLDD 180 HLNGWLESLF NDNEVKQKDT SCASGSGLRR DSQQWAFRRR NECSSCACGR HRGRRRGFQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 6e-23 | 9 | 110 | 4 | 104 | MYB PROTO-ONCOGENE PROTEIN |
| 1mse_C | 6e-23 | 9 | 110 | 4 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 6e-23 | 9 | 110 | 4 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator, when associated with BHLH002/EGL3/MYC146, BHLH012/MYC1, or BHLH042/TT8. {ECO:0000269|PubMed:15361138}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00213 | DAP | Transfer from AT1G66370 | Download |
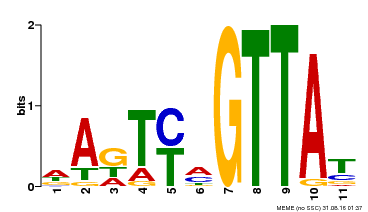
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.01G117100.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021632769.1 | 1e-167 | transcription factor MYB90-like | ||||
| Swissprot | Q9FNV9 | 1e-65 | MY113_ARATH; Transcription factor MYB113 | ||||
| TrEMBL | A0A2C9WK05 | 1e-177 | A0A2C9WK05_MANES; Uncharacterized protein | ||||
| STRING | POPTR_0005s14480.1 | 3e-75 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G66370.1 | 4e-68 | myb domain protein 113 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.01G117100.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



