 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.01G125000.3.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 797aa MW: 89172.2 Da PI: 6.4802 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 73.8 | 2.1e-23 | 127 | 228 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFk 83
f+k+lt sd++++g +++p++ ae+ +++++ ++l+ +d++ +W++++i+r++++r++lt+GW+ Fv+a++L +gD+v+F
Manes.01G125000.3.p 127 FCKTLTASDTSTHGGFSVPRRAAEKVfppldfsQQPPA--QELIARDLHDVEWKFRHIFRGQPKRHLLTTGWSVFVSAKRLVAGDSVLFI 214
99*****************************8544444..48************************************************ PP
E-SSSEE..EEEEE-S CS
B3 84 ldgrsefelvvkvfrk 99
++++ +l+++++r+
Manes.01G125000.3.p 215 --WNEKNQLLLGIRRA 228
..8899999****997 PP
| |||||||
| 2 | Auxin_resp | 120 | 1.9e-39 | 253 | 336 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+t+s F+v+YnPras+seFv++++k++ka+ ++vsvGmRf+m fete+ss rr++Gt++g+sdldpvrWpnS+Wrs+k
Manes.01G125000.3.p 253 AAHAAATNSCFTVFYNPRASPSEFVIPLSKYVKAVFhTRVSVGMRFRMLFETEESSVRRYMGTITGISDLDPVRWPNSHWRSVK 336
79********************************87599******************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 1.96E-49 | 111 | 256 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 6.7E-42 | 120 | 242 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 2.32E-21 | 126 | 227 | No hit | No description |
| PROSITE profile | PS50863 | 12.337 | 127 | 229 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.1E-23 | 127 | 229 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 1.2E-21 | 127 | 228 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 1.7E-34 | 253 | 336 | IPR010525 | Auxin response factor |
| SuperFamily | SSF54277 | 3.63E-7 | 685 | 765 | No hit | No description |
| Pfam | PF02309 | 2.0E-7 | 688 | 775 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 24.637 | 691 | 775 | IPR000270 | PB1 domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 797 aa Download sequence Send to blast |
MKLSTSGLGQ QGHEGEKKCL NSELWHACAG PLVSLPTVGS RVVYFPQGHS EQVAATTNKE 60 VDGHIPNYPS LPPQLICQLH NVTMHADVET DEVYAQMTLQ PLTPQEQKDT FLPMELGMPS 120 KQPTNYFCKT LTASDTSTHG GFSVPRRAAE KVFPPLDFSQ QPPAQELIAR DLHDVEWKFR 180 HIFRGQPKRH LLTTGWSVFV SAKRLVAGDS VLFIWNEKNQ LLLGIRRATR PQTVMPSSVL 240 SSDSMHIGLL AAAAHAAATN SCFTVFYNPR ASPSEFVIPL SKYVKAVFHT RVSVGMRFRM 300 LFETEESSVR RYMGTITGIS DLDPVRWPNS HWRSVKVGWD ESTAGERQPR VSLWEIEPLT 360 TFPMYPSLFP LRLRRPWHPG PSSLHDNRDE AGNGLMWLRG GTGEQGLHSL NFQAVNMFPW 420 AQQRLDPALL GNDQSQWYQA MLATGLQNVG SGDPLRQQFM QFQQPFQYLQ QSSSHCPLLQ 480 LQQQHQAIQQ STSHNVLQAQ NQISTESLPQ HLLQQQHNNQ PDDHAQQQQQ HHNYHDALQI 540 QASTTPIQNM LGSLCAEGSG NLLDFTRNGQ STLSEQLPQQ SWVQKYAHLQ VNAFTNSLSL 600 PRPYPEKVPA MEPENCNLDA QNATNFGMNI DSSGLLLPTT LPRYTSSTVD ADVSSMPLGD 660 SGFQNSIYGG MQDSSEILPS TGQVDPPTPS RTFVKVYKSG SVGRSLDISR FSSYHQLREE 720 LAQMFGIEGK LENPHRSGWQ LVFVDRENDV LLLGDDPWEA FVNNVWYIKI LSPEDVQKMG 780 EQGLESLGPN VGQRIE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 1e-161 | 16 | 357 | 47 | 388 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Regulates both stamen and gynoecium maturation. Promotes jasmonic acid production. Partially redundant with ARF6. Involved in fruit initiation. Acts as an inhibitor to stop further carpel development in the absence of fertilization and the generation of signals required to initiate fruit and seed development. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:16107481, ECO:0000269|PubMed:16829592}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00033 | PBM | Transfer from AT5G37020 | Download |
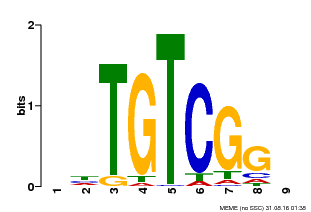
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.01G125000.3.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR167. {ECO:0000269|PubMed:17021043}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021592738.1 | 0.0 | auxin response factor 8-like isoform X2 | ||||
| Swissprot | Q9FGV1 | 0.0 | ARFH_ARATH; Auxin response factor 8 | ||||
| TrEMBL | A0A2C9WMH4 | 0.0 | A0A2C9WMH4_MANES; Auxin response factor | ||||
| STRING | cassava4.1_001923m | 0.0 | (Manihot esculenta) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G37020.2 | 0.0 | auxin response factor 8 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.01G125000.3.p |



