 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.02G118800.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | C3H | ||||||||
| Protein Properties | Length: 694aa MW: 74951.9 Da PI: 5.3073 | ||||||||
| Description | C3H family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-CCCH | 19.7 | 1.5e-06 | 263 | 283 | 5 | 26 |
--SGGGGTS--TTTTT-SS-SS CS
zf-CCCH 5 lCrffartGtCkyGdrCkFaHg 26
+C+ f++ G C+ Gd C +aHg
Manes.02G118800.2.p 263 PCPEFRK-GSCRQGDACEYAHG 283
8******.*************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF48403 | 1.49E-15 | 58 | 137 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.5E-16 | 59 | 138 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.0013 | 59 | 89 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50297 | 17.051 | 59 | 129 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 7.7E-10 | 59 | 128 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.603 | 59 | 83 | IPR002110 | Ankyrin repeat |
| CDD | cd00204 | 1.68E-14 | 61 | 134 | No hit | No description |
| PROSITE profile | PS50088 | 12.636 | 94 | 129 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 3.8E-4 | 94 | 126 | IPR002110 | Ankyrin repeat |
| SMART | SM00356 | 1.0E-4 | 258 | 284 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 12.662 | 263 | 285 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 52 | 293 | 316 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 6.118 | 293 | 317 | IPR000571 | Zinc finger, CCCH-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 694 aa Download sequence Send to blast |
MENEFQKRDG FFCDLSVLLE LSASNDLIGF KREIEGGRDI DERGLWYGRR IGSKRMGFEE 60 RTPLMIAALY GSKDVLNYIL EMGCVDVNRS CGSDGATALH CAAAGGSASS LEVVKLLLDA 120 SADPNTVDAD GNHASDLLVP FVGSDSNSRG KALELVLKGG CANDESCVVV DQNPNEMDGQ 180 DKQEVSTPIL SKDGTEKKEY PVDVTLPDIK NGIYGTDEFR MYTFKVKPCS RAYSHDWTEC 240 PFVHPGENAR RRDPRKYHYS CVPCPEFRKG SCRQGDACEY AHGIFECWLH PAQYRTRLCK 300 DEINCTRRVC FFAHKPEELR PLYASTGSAV PSPRSFSANG SALDIGSVSP LALGSPAVLI 360 PATSTPPLTP SGSSSPMGGW PNQSNIVPPT LHLPGSRLKS ALCARDMDLD MELLGLDNHH 420 RWQQQLMDEI SGLSSPSNWN NGLSTGSAFA VSGDRTAELN RIGGVKPTNL EDIFGSLDPS 480 ILPQLQGLSV DASASQLQSP TGIQMHQNIN QKLRSGYPTN FSSSPVRTSS FGIDPSGAAA 540 AAAAVLNSRA AAFAKRSQSF IERSAVNRHP GFSSPTSSAT VMPSNISDWG SPDGKLDWGI 600 QGEELNKLRK SASFGIRSNA SSLAAAAVSL PATLDEPDVS WVQSLVKDTS PLKSGHLGFE 660 EQQQQQCHTN TGSSEMLPAW MEQLYIEQEQ MVA* |
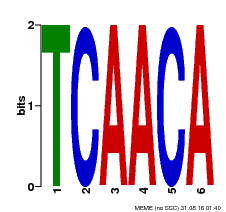
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00566 | DAP | Transfer from AT5G58620 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.02G118800.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021604126.1 | 0.0 | zinc finger CCCH domain-containing protein 66-like | ||||
| Refseq | XP_021604127.1 | 0.0 | zinc finger CCCH domain-containing protein 66-like | ||||
| Swissprot | Q9LUZ4 | 0.0 | C3H66_ARATH; Zinc finger CCCH domain-containing protein 66 | ||||
| TrEMBL | A0A2C9WD67 | 0.0 | A0A2C9WD67_MANES; Uncharacterized protein | ||||
| STRING | cassava4.1_002672m | 0.0 | (Manihot esculenta) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G58620.1 | 0.0 | C3H family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.02G118800.2.p |



