 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.03G137600.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 575aa MW: 62297.9 Da PI: 6.8541 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 101 | 7.2e-32 | 224 | 281 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
dDgynWrKYGqK+vkgsefprsYY+Ct+++C+vkk ers+ d++++ei+Y+g+H+h+k
Manes.03G137600.1.p 224 DDGYNWRKYGQKHVKGSEFPRSYYKCTHPNCEVKKLFERSH-DGQITEIVYKGTHDHPK 281
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 106.2 | 1.6e-33 | 389 | 447 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+v+g+++prsYY+Ct agCpv+k+ver+++dpk+v++tYeg+Hnh+
Manes.03G137600.1.p 389 LDDGYRWRKYGQKVVRGNPNPRSYYKCTNAGCPVRKHVERASHDPKAVITTYEGKHNHD 447
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 3.3E-27 | 221 | 282 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 8.5E-25 | 222 | 282 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.8E-34 | 223 | 281 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.3E-24 | 224 | 280 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 22.905 | 224 | 282 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.3E-37 | 374 | 449 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.22E-29 | 381 | 449 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.825 | 384 | 449 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.1E-39 | 389 | 448 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.4E-26 | 390 | 447 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 575 aa Download sequence Send to blast |
MDNSSSESEL QLCGPSHGVS DSGDPMRHES DAAGVAGSGG ARYKLMSPAK LPISRSPCIT 60 IPPGLSPTSF LESPVLLSNV KVEPSPTTGS LTKPQMEHGF LGSNTYSMKV PSNALEERKY 120 SCFEFRPHAR SNLVPADMNH QRSECVQGQC QSQSHPPSPA VKNEMAVPSN EFSLTAPLPT 180 VTSGVSAPTE IDSDELNQMG ISNSGLQASQ SDHKGGNGIS MPSDDGYNWR KYGQKHVKGS 240 EFPRSYYKCT HPNCEVKKLF ERSHDGQITE IVYKGTHDHP KPQPSRRYAA GAVLSMQEDR 300 SDKISSLPGG DDKSSSAFGQ VPNTIEPNNA PELSPVMTND DSIEGAEDED DSFSKRRKMD 360 TGGFDVTPVI KPIREPRVVV QTLSEVDILD DGYRWRKYGQ KVVRGNPNPR SYYKCTNAGC 420 PVRKHVERAS HDPKAVITTY EGKHNHDVPT ARSSSHDTVG PTLVNGSSRI RSDENETISL 480 DLGVGISSTA ENRSTDQQHG PHSELTKSQT QTSGSGFRIV PRIPIAPSYS VLNGGVNQYG 540 SRQNPNEGRS VEIPALSHSS YPYPQNMGRL LTGP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-36 | 224 | 449 | 8 | 77 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-36 | 224 | 449 | 8 | 77 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. {ECO:0000250|UniProtKB:Q9SI37}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
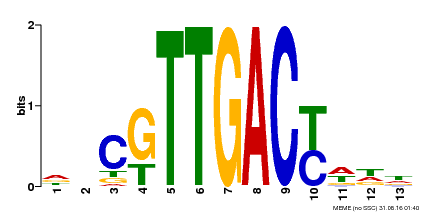
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.03G137600.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT827607 | 0.0 | KT827607.1 Manihot esculenta WRKY transcription factor 32 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021607976.1 | 0.0 | probable WRKY transcription factor 20 | ||||
| Swissprot | Q93WV0 | 0.0 | WRK20_ARATH; Probable WRKY transcription factor 20 | ||||
| TrEMBL | A0A2C9W7B0 | 0.0 | A0A2C9W7B0_MANES; Uncharacterized protein | ||||
| STRING | cassava4.1_004461m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3946 | 34 | 63 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.2 | 0.0 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.03G137600.1.p |



