 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.03G154500.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 262aa MW: 28957.7 Da PI: 7.8017 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 61.2 | 1.3e-19 | 182 | 216 | 1 | 35 |
GATA 1 CsnCgttkTplWRrgpdgnktLCnaCGlyyrkkgl 35
C++Cg+ kTp+WR gp g+ktLCnaCG++y++ +l
Manes.03G154500.1.p 182 CQHCGAEKTPQWRAGPLGPKTLCNACGVRYKSGRL 216
*******************************9885 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00401 | 1.0E-15 | 176 | 226 | IPR000679 | Zinc finger, GATA-type |
| PROSITE profile | PS50114 | 12.374 | 176 | 212 | IPR000679 | Zinc finger, GATA-type |
| Gene3D | G3DSA:3.30.50.10 | 8.0E-15 | 177 | 214 | IPR013088 | Zinc finger, NHR/GATA-type |
| SuperFamily | SSF57716 | 2.61E-15 | 178 | 240 | No hit | No description |
| CDD | cd00202 | 1.52E-15 | 181 | 229 | No hit | No description |
| Pfam | PF00320 | 3.7E-17 | 182 | 216 | IPR000679 | Zinc finger, GATA-type |
| PROSITE pattern | PS00344 | 0 | 182 | 207 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 262 aa Download sequence Send to blast |
MESLDPAACF IMDDLLDFAS DIGERDDDEH SNKPTKAFPP LNPSPNGLAV APLPFDVFDH 60 PDPSPEFAEE ELEWLSNKDA FPAVETFVDI ISENPGGLPK QRSPVSVLEN STTSSTSNSG 120 NSGTNGSITM DYCWSLQVPV KARSKHHRSR RRDLQGQQCW WSLENLRKVK PAVTSSTMGR 180 KCQHCGAEKT PQWRAGPLGP KTLCNACGVR YKSGRLVPEY RPASSPTFRS ELHSNSHRKV 240 MEMRKQKLIM GSMVVKAMEK G* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. May be involved in the regulation of some light-responsive genes. {ECO:0000269|PubMed:12139008}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00377 | DAP | Transfer from AT3G24050 | Download |
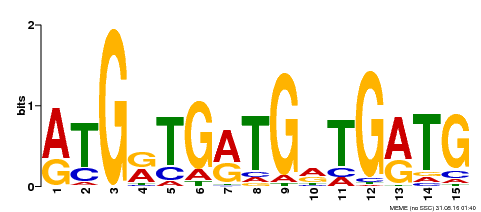
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.03G154500.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021607544.1 | 0.0 | GATA transcription factor 1-like isoform X1 | ||||
| Refseq | XP_021607545.1 | 0.0 | GATA transcription factor 1-like isoform X2 | ||||
| Swissprot | Q8LAU9 | 7e-54 | GATA1_ARATH; GATA transcription factor 1 | ||||
| TrEMBL | A0A251LB17 | 0.0 | A0A251LB17_MANES; Uncharacterized protein | ||||
| STRING | cassava4.1_014204m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8537 | 29 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24050.1 | 2e-53 | GATA transcription factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.03G154500.1.p |



