 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.05G022000.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 612aa MW: 67097.2 Da PI: 6.4597 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 40.5 | 5.1e-13 | 442 | 488 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI++Lq
Manes.05G022000.1.p 442 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAIAYINELQ 488
799***********************66......******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 1.1E-55 | 52 | 240 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| SuperFamily | SSF47459 | 3.93E-18 | 437 | 501 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.047 | 438 | 487 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.33E-15 | 441 | 492 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 5.8E-18 | 442 | 501 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.8E-10 | 442 | 488 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.3E-16 | 444 | 493 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 612 aa Download sequence Send to blast |
MKIEIGMGGG AWNDEDRAMV AAVLGTNALN YLISNSVSNE NLLMAIGSDE NLQNKLSDLV 60 DNPNAWNFSW NYAIFWQISC SKSGDCVLGW GDGSCREPKE GEESEGTRIL NLRFQDETQQ 120 RMRKRVLQKL HTLSGESDDD NYALGLDRVT DTEMFFLASM YFSFASGEGG PGKCLASGKH 180 VWIADALKSG NDYCVRSFLA KSAGIQTIVL VPTDAGVVEL GSVRSVYESM DIVQSIRSTF 240 STNSTVIRAK SMVPVAAPAL PVVNTKKDDN SLFSNVGIVE RVEGIPKIFG QELNHGHGQG 300 YREKLAVRKM EERPSWDVYQ NGNRLAFSGN RNGLHGSSWP HGFGLKQGSP VEVYGSQATT 360 NNIQDLVNGA RNQFQPQKQV QMQIDFSGAT SGPSVIGLPV SVESEHSDVE ASCKEERPGT 420 AEDRRPRKRG RKPANGREEP LNHVEAERQR REKLNQRFYA LRAVVPNISK MDKASLLGDA 480 IAYINELQAK LKLMEAEREK FGSTSRESSA LEVNSNGKNH CQTSEVDIQA SHDEVIVRVS 540 CPLDSHPASR VIKAFKEAQV SVIDSKLAAA NDTVFHTFVI KSQGSEQLTK EKLMAAFSHE 600 SNSLQQVSSV G* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 2e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 2e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 2e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 2e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 2e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 2e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 2e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 2e-29 | 436 | 498 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 423 | 431 | RRPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that negatively regulates jasmonate (JA) signaling (PubMed:30610166). Negatively regulates JA-dependent response to wounding, JA-induced expression of defense genes, JA-dependent responses against herbivorous insects, and JA-dependent resistance against Botrytis cinerea infection (PubMed:30610166). Plays a positive role in resistance against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (PubMed:30610166). {ECO:0000269|PubMed:30610166}. | |||||
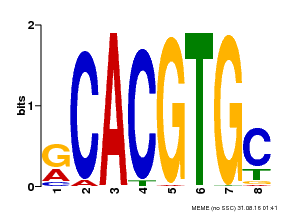
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.05G022000.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding, feeding with herbivorous insects, infection with the fungal pathogen Botrytis cinerea and infection with the bacterial pathogen Pseudomonas syringae pv tomato DC3000. {ECO:0000269|PubMed:30610166}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021613223.1 | 0.0 | transcription factor bHLH13-like | ||||
| Refseq | XP_021613224.1 | 0.0 | transcription factor bHLH13-like | ||||
| Swissprot | A0A3Q7ELQ2 | 0.0 | MTB1_SOLLC; Transcription factor MTB1 | ||||
| TrEMBL | A0A2C9VUS1 | 0.0 | A0A2C9VUS1_MANES; Uncharacterized protein | ||||
| STRING | cassava4.1_003856m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5448 | 32 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 0.0 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.05G022000.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



