 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.05G039400.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 823aa MW: 89065.2 Da PI: 6.0325 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66.1 | 4.6e-21 | 116 | 171 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
Manes.05G039400.1.p 116 KKRYHRHTPQQIQELEALFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 171
688999***********************************************999 PP
| |||||||
| 2 | START | 210.8 | 5.1e-66 | 326 | 550 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S.... CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla.... 77
ela++a++elvk+a+ +ep+W +s e++n +e++++f++ + + +ea+r++g+v+ ++ lve+l+d++ +W e+++
Manes.05G039400.1.p 326 ELALAAMDELVKMAQTDEPLWIRSLeggrEILNHEEYMRTFTPCIGmkpggFVSEASRETGMVIINSLALVETLMDSN-RWAEMFPcmia 414
5899**************************************999899******************************.*********** PP
EEEEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTC CS
START 78 kaetlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksng 160
+ +t++vis+g g lqlm aelq+lsplvp R++ f+R+++q+ +g+w++vdvS+d ++ + + +v +++lpSg+++++++ng
Manes.05G039400.1.p 415 RTSTTDVISNGmggtrnGSLQLMLAELQVLSPLVPvREVNFLRFCKQHAEGVWAVVDVSIDTIRETSGAPAFVNCRRLPSGCVVQDMPNG 504
*****************************************************************999********************** PP
EEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 161 hskvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
+skvtwveh+++++ ++h+l+r+l++sg+ +ga++wvatlqrqce+
Manes.05G039400.1.p 505 YSKVTWVEHAEYDETQIHQLYRPLISSGMGFGAQRWVATLQRQCEC 550
********************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.07E-20 | 99 | 173 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 9.2E-22 | 103 | 173 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.216 | 113 | 173 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 9.9E-18 | 114 | 177 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.33E-18 | 115 | 173 | No hit | No description |
| Pfam | PF00046 | 1.4E-18 | 116 | 171 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 148 | 171 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 44.819 | 317 | 553 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 6.87E-34 | 319 | 550 | No hit | No description |
| CDD | cd08875 | 1.00E-127 | 321 | 549 | No hit | No description |
| Pfam | PF01852 | 1.4E-57 | 326 | 550 | IPR002913 | START domain |
| SMART | SM00234 | 3.7E-51 | 326 | 550 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.35E-23 | 578 | 748 | No hit | No description |
| SuperFamily | SSF55961 | 2.35E-23 | 775 | 815 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 823 aa Download sequence Send to blast |
MSFGGFLENG SPGGGGARIV ADIPYSSSNM PTGAIAQPRL ISPSLTKAMF NSPGLSLALQ 60 QPNIDGQGDI ARMAENFESN GGRRSREEEH ESRSGSDNMD GASGDDQDAA DNPPRKKRYH 120 RHTPQQIQEL EALFKECPHP DEKQRLELSK RLCLETRQVK FWFQNRRTQM KTQLERHENS 180 LLRQENDKLR AENMSIRDAM RNPICSNCGG PAIIGDISLE EQHLRIENAR LKDELDRVCA 240 LAGKFLGRPI SSLAGSIGPP MPNSSLELGV GTNGFSGLST VPATLPLGPD FAGGISGALP 300 VMTQTRPATA GVTGLDRSFE RSMFLELALA AMDELVKMAQ TDEPLWIRSL EGGREILNHE 360 EYMRTFTPCI GMKPGGFVSE ASRETGMVII NSLALVETLM DSNRWAEMFP CMIARTSTTD 420 VISNGMGGTR NGSLQLMLAE LQVLSPLVPV REVNFLRFCK QHAEGVWAVV DVSIDTIRET 480 SGAPAFVNCR RLPSGCVVQD MPNGYSKVTW VEHAEYDETQ IHQLYRPLIS SGMGFGAQRW 540 VATLQRQCEC LAILMSSAVP TRDHTAITAS GRRSMLKLAQ RMTDNFCAGV CASTVHKWNK 600 LNAGNVDEDV RVMTRKSVDD PGEPPGIVLS AATSVWLPVS PQRLFDFLRD ERLRSEWDIL 660 SNGGPMQEMA HIAKGQDHGN CVSLLRASAM NANQSSMLIL QETCIDAAGS LVVYAPVDIP 720 AMHVVMNGGD SAYVALLPSG FAIVPDGPGS RGSLSTPNGP TGNNGGGTGG QQRVSGSLLT 780 VAFQILVNSL PTAKLTVESV ETVNNLISCT VQKIKAALQC ES* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
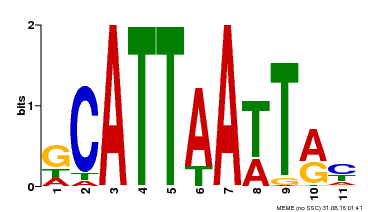
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.05G039400.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021612685.1 | 0.0 | homeobox-leucine zipper protein ANTHOCYANINLESS 2-like | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A2C9VVI4 | 0.0 | A0A2C9VVI4_MANES; Uncharacterized protein | ||||
| STRING | cassava4.1_001764m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1192 | 34 | 106 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.05G039400.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



