 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.05G106900.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 525aa MW: 57166 Da PI: 8.7663 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 108.4 | 3.5e-34 | 241 | 297 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
dDgynWrKYGqK+vkgsefprsYY+Ct+++Cpvkkkvers d++v+ei+Y+g+Hnh+
Manes.05G106900.1.p 241 DDGYNWRKYGQKQVKGSEFPRSYYKCTHPSCPVKKKVERSL-DGQVTEIIYKGQHNHQ 297
8****************************************.***************8 PP
| |||||||
| 2 | WRKY | 106.2 | 1.7e-33 | 412 | 470 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct++gC+v+k+ver+a+dp++v++tYeg+Hnh+
Manes.05G106900.1.p 412 LDDGYRWRKYGQKVVKGNPYPRSYYKCTTSGCTVRKHVERAATDPRAVITTYEGKHNHD 470
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 2.8E-29 | 237 | 299 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.53E-26 | 238 | 299 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.6E-37 | 240 | 298 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.709 | 240 | 299 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.3E-26 | 241 | 297 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 8.7E-37 | 397 | 472 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.44E-29 | 404 | 472 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.595 | 407 | 472 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.7E-38 | 412 | 471 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.6E-26 | 413 | 470 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 525 aa Download sequence Send to blast |
MADNGTRHPS PPVSSPPPTS KSSSTSSARP ATISLPPRSF NETFFTSGIG MGFSPGPMTL 60 VSSFFSDSDD FKSFSQLLAG AMASPAANTN NNKLPFPPPQ QSKNSLADVS KPANLSIVPP 120 SPMFNLPPGL SPMALLDSPG FGLLSPQGSF GMTHQQALAQ VTAQAVHAHS SMHIQAQYSS 180 SLPSAQATSS TQFSSVTTNS TTLQQMLPSI PDPSDSIKES SDFTHSDQRS QAYSLGDKPN 240 DDGYNWRKYG QKQVKGSEFP RSYYKCTHPS CPVKKKVERS LDGQVTEIIY KGQHNHQPPQ 300 PNKRVKDAGS LKGNSDNQNN SELASQIQVG KMNKSKDRKD QESSQATPEL VSGTSDSEEV 360 GDTETAVDEN DEDEPNPKRR NTEVKVTEPA SSHRTVTEPR IIVQTTSEVD LLDDGYRWRK 420 YGQKVVKGNP YPRSYYKCTT SGCTVRKHVE RAATDPRAVI TTYEGKHNHD VPAAKGSSHG 480 TTSSNPSERK QQNVEKHSLD NRRDFGTNNQ QPIARLRLKE EQIT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 8e-43 | 241 | 473 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 8e-43 | 241 | 473 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00260 | DAP | Transfer from AT2G03340 | Download |
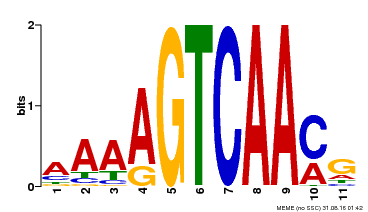
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.05G106900.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid and during leaf senescence. {ECO:0000269|PubMed:11449049, ECO:0000269|PubMed:11722756}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT827601 | 0.0 | KT827601.1 Manihot esculenta WRKY transcription factor 26 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021613186.1 | 0.0 | probable WRKY transcription factor 3 | ||||
| Swissprot | Q9ZQ70 | 0.0 | WRKY3_ARATH; Probable WRKY transcription factor 3 | ||||
| TrEMBL | A0A2C9VWJ7 | 0.0 | A0A2C9VWJ7_MANES; Uncharacterized protein | ||||
| STRING | cassava4.1_006823m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1545 | 34 | 100 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G03340.1 | 1e-159 | WRKY DNA-binding protein 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.05G106900.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



