 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.09G181800.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 736aa MW: 78256.6 Da PI: 6.3694 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.6 | 3.3e-16 | 467 | 513 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
Manes.09G181800.1.p 467 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 513
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.62E-17 | 459 | 517 | No hit | No description |
| SuperFamily | SSF47459 | 1.57E-20 | 460 | 524 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.8E-20 | 460 | 521 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.349 | 463 | 512 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.1E-13 | 467 | 513 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.7E-17 | 469 | 518 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 736 aa Download sequence Send to blast |
MPLSELVYRL AKGKLDSSQE KNPTCSTDLS SEPETDFVEL VWENGQIQSS KTRKIQFSSS 60 FPSQTSKIRD KDVANGSNTK MGRFGAMDSV IGEVPMSVPS VEMGLNQDDD MVPWLNYPIE 120 DSLQHDYCSE FLAELSGVTV NENSQTNYAS MEKRNCGNQS VRDPCPASVR NSLSLEQGHI 180 SRVSSTGDVD ASRPRTSSSQ LYPSPSQQCQ TSFPYFRSRV SAGNSESMSN LARHAASGDS 240 IGVPPSGGGV PSAKMQKQVP VSSATNPSLM NFSHFSRPAA LVKANLCNMG TKAGSGISNM 300 EKMVSKDKGS IASRSNPAES ILADTFSGLR KDISSNCHPA MVLPKVDAKS LDPTPAEESL 360 PAKRPEALDQ EDCKSDKNHC QLAESTTRGL VDGEKTAEHL VASSSVCSGN SVERVSDERT 420 QDLKRKHRET EDSEGPSEDV EEESVGAKKA APARGGPGSK RSRAAEVHNL SERRRRDRIN 480 EKMRALQELI PNCNKVDKAS MLDEAIEYLK TLQLQVQIMS MGAGLYMPSM MLPPGMPHMH 540 AGHMVQFSPM GVGMGMGMGM GYGMGMPDMI GGSSGCSMIQ APPMHGAHFP GPPMSGPSAL 600 HGMGGSNLPI FGLSAQGHPM PYPCAPLMPV SGGPLLKTTM GLNAGGLAGP VDNLNSGPGS 660 RSKDAIQNIS QAMQNSGANS SMNQTSTQCQ VTNERFEQHP SVQNSGQASE VTDRGAVKSA 720 NGNDNTPNQV TAGCD* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 471 | 476 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
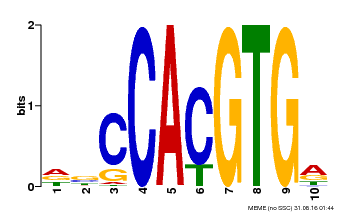
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.09G181800.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021623245.1 | 0.0 | transcription factor PIF3-like isoform X1 | ||||
| Refseq | XP_021623246.1 | 0.0 | transcription factor PIF3-like isoform X1 | ||||
| TrEMBL | A0A2C9VE55 | 0.0 | A0A2C9VE55_MANES; Uncharacterized protein | ||||
| STRING | cassava4.1_002400m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5271 | 33 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 1e-52 | phytochrome interacting factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.09G181800.1.p |



