 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.13G081100.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 308aa MW: 33107 Da PI: 8.7166 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 66.8 | 4.2e-21 | 112 | 161 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrd.psengkr.krfslgkfgtaeeAakaaiaarkkleg 55
+y+GVr+++ +g+W+AeIrd + + r++lg+f+tae Aa+a+++a+++++g
Manes.13G081100.2.p 112 KYRGVRQRP-WGKWAAEIRDpF-----KaSRVWLGTFDTAEAAARAYDEAALRFRG 161
8********.**********66.....56************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.730.10 | 1.9E-33 | 111 | 170 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 5.04E-23 | 112 | 170 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 1.23E-23 | 112 | 171 | No hit | No description |
| PROSITE profile | PS51032 | 25.673 | 112 | 169 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.6E-38 | 112 | 175 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.1E-14 | 112 | 161 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.0E-12 | 113 | 124 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.0E-12 | 135 | 151 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006970 | Biological Process | response to osmotic stress | ||||
| GO:0009749 | Biological Process | response to glucose | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 308 aa Download sequence Send to blast |
MASELCRERE MSAMVSALIH VVTGKIPKPS SSEYSLCNEG ANCSSSSSAG GGAKRPREAD 60 NGCQDFTKLS RPLVDDEFSH GRAATTIIPP AEPMFTAIYE HNNTFQEEPM RKYRGVRQRP 120 WGKWAAEIRD PFKASRVWLG TFDTAEAAAR AYDEAALRFR GSKAKLNFPE NVKLRPSTPN 180 AHLMNSDSPN TLLSVPTSTQ VPMVNSQVAG SGESVNHSQL VLGIGSYQRP AMNLYDKMVL 240 PSSTASLHSQ SSTFVGSTSS TFLLSSSLPH FFPPPSLGYL RRATNQSGGA DTSGPSWSGS 300 SHCSPSP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 2e-23 | 111 | 172 | 1 | 63 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00584 | DAP | Transfer from AT5G64750 | Download |
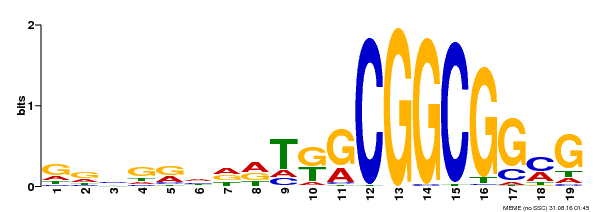
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.13G081100.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021633133.1 | 0.0 | ethylene-responsive transcription factor ABR1-like, partial | ||||
| TrEMBL | A0A2C9UR30 | 0.0 | A0A2C9UR30_MANES; Uncharacterized protein | ||||
| STRING | cassava4.1_026922m | 0.0 | (Manihot esculenta) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64750.1 | 2e-26 | ERF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.13G081100.2.p |



