 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.14G098000.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 367aa MW: 40348.4 Da PI: 9.6623 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 43.8 | 6.4e-14 | 66 | 114 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ ++r++lg+f ++eAa+a++ a+++++g
Manes.14G098000.1.p 66 SKYKGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNEEDEAARAYDIAAQRFRG 114
89****9888.8*********3.....5**********99*************98 PP
| |||||||
| 2 | B3 | 105.1 | 3.5e-33 | 199 | 299 | 1 | 96 |
EEEE-..-HHHHTT-EE--HHH.HTT.....---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE- CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.....ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkld 85
f+k+ tpsdv+k++rlv+pk++ae+h g+++++++ l +ed sg++W+++++y+++s++yvltkGW++Fvk+++Lk+gD+v F+++
Manes.14G098000.1.p 199 FEKAVTPSDVGKLNRLVIPKQHAEKHfplqsGNNSTKGVLLNFEDISGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLKAGDIVSFQRS 288
89*****************************99999****************************************************87 PP
SSSEE..EEEE CS
B3 86 grsefelvvkv 96
+++l++ +
Manes.14G098000.1.p 289 TGPDKQLYIDW 299
66777777766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.14E-25 | 66 | 124 | No hit | No description |
| Pfam | PF00847 | 7.1E-9 | 66 | 114 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 3.27E-17 | 66 | 122 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 4.2E-28 | 67 | 128 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 20.297 | 67 | 122 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.4E-20 | 67 | 122 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 2.4E-42 | 193 | 305 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 3.79E-32 | 197 | 300 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 8.49E-30 | 198 | 288 | No hit | No description |
| PROSITE profile | PS50863 | 14.565 | 199 | 303 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 4.7E-30 | 199 | 300 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 2.6E-25 | 199 | 303 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 367 aa Download sequence Send to blast |
MDGSCTDEST TSETLSISIS PTSAVSSFPP PTKSPESPLG RVGSGSSVIL DSESGVEAES 60 RKLPSSKYKG VVPQPNGRWG AQIYEKHQRV WLGTFNEEDE AARAYDIAAQ RFRGRDAITN 120 FKPQGAETEA DDIEAAFLNS HSKAEIVDML RKHTYDDELE QSKRNYKIDG LGKQNRNPGA 180 NNVALSGSDR VLKAREQLFE KAVTPSDVGK LNRLVIPKQH AEKHFPLQSG NNSTKGVLLN 240 FEDISGKVWR FRYSYWNSSQ SYVLTKGWSR FVKEKNLKAG DIVSFQRSTG PDKQLYIDWK 300 ARNGSNPVVC SVQPVQMVRL FGVNIFKAPG GSGVAEGAGG CNGKRMREME LLSLNCIKKQ 360 RIIGAL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 7e-63 | 196 | 306 | 11 | 121 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds specifically to bipartite recognition sequences composed of two unrelated motifs, 5'-CAACA-3' and 5'-CACCTG-3'. May function as negative regulator of plant growth and development. {ECO:0000269|PubMed:15040885, ECO:0000269|PubMed:9862967}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00139 | DAP | Transfer from AT1G13260 | Download |
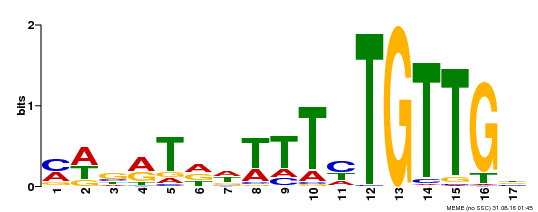
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.14G098000.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by brassinosteroid and zeatin. {ECO:0000269|PubMed:15040885}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021592563.1 | 0.0 | AP2/ERF and B3 domain-containing transcription factor RAV1 | ||||
| Swissprot | Q9ZWM9 | 1e-155 | RAV1_ARATH; AP2/ERF and B3 domain-containing transcription factor RAV1 | ||||
| TrEMBL | A0A2C9UJZ4 | 0.0 | A0A2C9UJZ4_MANES; Uncharacterized protein | ||||
| STRING | cassava4.1_010157m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2920 | 32 | 74 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25560.1 | 1e-152 | RAV family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.14G098000.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



