 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.15G063600.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 580aa MW: 62997.3 Da PI: 6.9757 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 101 | 6.9e-32 | 222 | 279 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
dDgynWrKYGqK+vkgsefprsYY+Ct+++C+vkk ers+ d++++ei+Y+g+H+h+k
Manes.15G063600.1.p 222 DDGYNWRKYGQKHVKGSEFPRSYYKCTHPNCEVKKLFERSH-DGQITEIIYKGTHDHPK 279
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 106.2 | 1.7e-33 | 386 | 444 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+v+g+++prsYY+Ct agCpv+k+ver+++dpk+v++tYeg+Hnh+
Manes.15G063600.1.p 386 LDDGYRWRKYGQKVVRGNPNPRSYYKCTNAGCPVRKHVERASHDPKAVITTYEGKHNHD 444
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 6.9E-27 | 220 | 280 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.29E-24 | 221 | 280 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.1E-34 | 221 | 279 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.7E-24 | 222 | 278 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 22.791 | 222 | 280 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.3E-37 | 371 | 446 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.35E-29 | 378 | 446 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.825 | 381 | 446 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.1E-39 | 386 | 445 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.5E-26 | 387 | 444 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 580 aa Download sequence Send to blast |
MDNTSNESEL RLCGASDPGD PTLRESDAAG AASSGGARYK LMSPAKLPIS RSPCITIPPG 60 LSPTSFLESP VLLSNVKAEP SPTTGSFTNT QTGHGFLGSN SYSVMAPSNA YGERKSSCFE 120 CRPHTRSNLV PADVNHQRTE QSVQVQGQYY SQSHASSPTV KSEEVPLNEL SLSAPPPMVT 180 SGSSAPTEVD SEELNQMGAS NSGLQVSQSD HKGGSGLSMS SDDGYNWRKY GQKHVKGSEF 240 PRSYYKCTHP NCEVKKLFER SHDGQITEII YKGTHDHPKP QPSRRYTAGA VLLMQEDRSD 300 KISLHGRDDK SPSAYGQVSN TIEPNGTPEL SPLTPNDDGT EGAEDDYDPF SKRRKMDTGG 360 FDVTPVIKPI REPRVVVQTL SEVDILDDGY RWRKYGQKVV RGNPNPRSYY KCTNAGCPVR 420 KHVERASHDP KAVITTYEGK HNHDVPTARS SSHDTARSSS HDTAGPTPVN GQSRIRSDES 480 DTISLDLGVG ICSTAENRSN DQQQAMHSEF TQNRNQTSGS SFRIAPRTPI TPYYGVLNGG 540 MHQFGSRQNP NEGHSVEIPP LSHSSCPYPQ NMGRLVMGP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 2e-36 | 213 | 446 | 4 | 77 | Probable WRKY transcription factor 4 |
| 2lex_A | 2e-36 | 213 | 446 | 4 | 77 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. {ECO:0000250|UniProtKB:Q9SI37}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
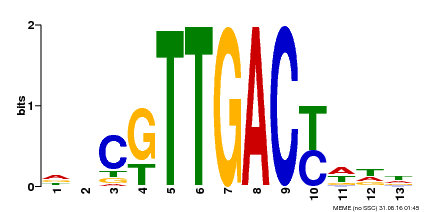
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.15G063600.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT827607 | 0.0 | KT827607.1 Manihot esculenta WRKY transcription factor 32 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021595661.1 | 0.0 | probable WRKY transcription factor 20 | ||||
| Swissprot | Q93WV0 | 0.0 | WRK20_ARATH; Probable WRKY transcription factor 20 | ||||
| TrEMBL | A0A140H8N6 | 0.0 | A0A140H8N6_MANES; WRKY transcription factor 32 | ||||
| STRING | cassava4.1_004372m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3946 | 34 | 63 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.2 | 1e-180 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.15G063600.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



