 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Manes.18G015400.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Euphorbiaceae; Crotonoideae; Manihoteae; Manihot
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 954aa MW: 104918 Da PI: 5.3516 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 76.3 | 3.5e-24 | 156 | 257 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFk 83
f+k+lt sd++++g +++p++ ae+ ++++ + ++l+++d++ ++W++++iyr++++r++lt+GW+ Fv ++Lk+gD+v+F
Manes.18G015400.1.p 156 FCKTLTASDTSTHGGFSVPRRAAEKLfpaldytMQPP-T-QELVVRDLHDNTWTFRHIYRGQPKRHLLTTGWSIFVGTKRLKAGDSVLFI 243
99*********************999*****954444.4.38************************************************ PP
E-SSSEE..EEEEE-S CS
B3 84 ldgrsefelvvkvfrk 99
++++ +l+v+v+r+
Manes.18G015400.1.p 244 --RDEKSQLLVGVRRA 257
..4577778*****97 PP
| |||||||
| 2 | Auxin_resp | 114.7 | 8.1e-38 | 282 | 365 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+++s+F+++YnPra++seFv++++k +ka++ ++vsvGmRf m+fete+s +rr++Gt+vg+sd+dp+rWp+SkWr+L+
Manes.18G015400.1.p 282 AAHAAANRSPFTIFYNPRACPSEFVIPLAKHRKAVYgTQVSVGMRFGMMFETEESGKRRYMGTIVGISDIDPLRWPGSKWRNLQ 365
79**********************************9*********************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 1.19E-46 | 143 | 285 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 2.3E-41 | 149 | 270 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.90E-22 | 155 | 256 | No hit | No description |
| Pfam | PF02362 | 8.3E-22 | 156 | 257 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.1E-23 | 156 | 258 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.042 | 156 | 258 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 5.4E-33 | 282 | 365 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 7.6E-10 | 831 | 926 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 26.393 | 839 | 923 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 2.4E-8 | 852 | 918 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0010305 | Biological Process | leaf vascular tissue pattern formation | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048507 | Biological Process | meristem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 954 aa Download sequence Send to blast |
MHFKQKHMMM GSVEEKIKAG GLVGGIQTNL LEEMKLLKEI QDHSGTRKTI NSELWYACAG 60 PLVSLPQVGS LVYYFPQGHS EQVAVSTKRT ATSQIPNYPN LPSQLLCQVH NVTLHADKDT 120 DEIYAQMSLQ PVNSEKDVFP IPDFGLKPSK HPTEFFCKTL TASDTSTHGG FSVPRRAAEK 180 LFPALDYTMQ PPTQELVVRD LHDNTWTFRH IYRGQPKRHL LTTGWSIFVG TKRLKAGDSV 240 LFIRDEKSQL LVGVRRANRQ QTTLPSSVLS ADSMHIGVLA AAAHAAANRS PFTIFYNPRA 300 CPSEFVIPLA KHRKAVYGTQ VSVGMRFGMM FETEESGKRR YMGTIVGISD IDPLRWPGSK 360 WRNLQVEWDE PGCSDKQNRV SPWEIETPES LFIFPSLTSG LKRPLHSGFL GGETEWGNLI 420 KRPLIWLPEN GNGNFPYSTI PNTCSERLLK MLMKPHDNHP GIYESALPEI AASKGTPLDD 480 MKAMQGTVNQ MPQLNQSVAM SVENQTYSQF CASQSNAMIS SSSKINLPGK LHLPCNLENQ 540 TADGISNEKL KSEPDHSTDK LSQVTSVGES NEEKSSSSPT NLQNCGNQMD FQNQNQAQLH 600 AQSSLWSVQP LLEPSGVHPQ QIHISQADSI PLSGSLPFLD TDEWISNPSC ISLPGMYGSS 660 GPLSMFGLQE QSAGLPDPSI PLMNQGLWDQ QLNNLRFLSP GSHLIPLAPQ EPCNLNSSGA 720 KDLSDESNDQ SGIYDTLNID VGNGGSAVID PSVSNVILDE FCTSKGVDFQ NPSDCLVGNF 780 SSSQDVQSQI TSASLADSQA FSQQEFPDSL GGTPSSNVDF EKGNYMQNNS WQQVAPRVRT 840 YTKVQKAGSV GRSIDVSSFR NYEELCSAIE CMFGLEGLLN NPRESGWKLV YVDYENDVLL 900 IGDDPWEEFV GCVRCIRILS PSEVQQMSEE GMKLLNNANI QGLAASVTEG SRA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 0.0 | 9 | 390 | 1 | 392 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Mediates embryo axis formation and vascular tissues differentiation. Functionally redundant with ARF7. May be necessary to counteract AMP1 activity. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:14973283, ECO:0000269|PubMed:17553903}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00153 | DAP | Transfer from AT1G19850 | Download |
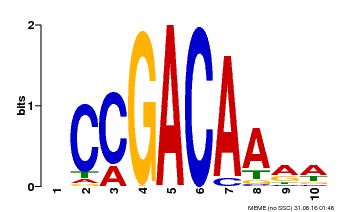
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Manes.18G015400.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021600166.1 | 0.0 | auxin response factor 5 | ||||
| Swissprot | P93024 | 0.0 | ARFE_ARATH; Auxin response factor 5 | ||||
| TrEMBL | A0A2C9U0H6 | 0.0 | A0A2C9U0H6_MANES; Auxin response factor | ||||
| STRING | cassava4.1_001105m | 0.0 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8991 | 32 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19850.1 | 0.0 | ARF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Manes.18G015400.1.p |



