 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0001s0298.3.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 1111aa MW: 119341 Da PI: 8.2272 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 119.2 | 5.1e-37 | 588 | 752 | 2 | 128 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgt.......ssssa..sec.eaesssss 81
g+kdrhsk+ T++g+RdRRvRls+++a +f+d+qd+LGfd++sk++eWL+ +ak ai+el + + s c ++ ++++s
Mapoly0001s0298.3.p 588 SGGKDRHSKVNTAKGPRDRRVRLSVPTAVQFYDVQDRLGFDQPSKAVEWLIHKAKDAIDELGRIpvdgkdgP---RvpSVCtSDITRHVS 674
689************************************************************966665531...144556544444444 PP
TCP 82 asnsssg.................................kaaksaakskksqksaasalnlak.esrakarararertre 128
s+s g + ++ +a++ +s++sa + la+ esr kar+rarert++
Mapoly0001s0298.3.p 675 ISSSY-GagavsglgmfaplqpnqgnfagptfgvgrmdltRPNSGSAEGGRSNESAG--SPLARvESRTKARERARERTKK 752
44222.13556677788888888889999********98655555556666666666..899999**************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 9.9E-35 | 589 | 752 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 32.971 | 590 | 648 | IPR017887 | Transcription factor TCP subgroup |
| PROSITE profile | PS51370 | 9.434 | 736 | 753 | IPR017888 | CYC/TB1, R domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0045962 | Biological Process | positive regulation of development, heterochronic | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1111 aa Download sequence Send to blast |
MLAWARSVDS STSCHARSTS WRIGFEGNCW PVERDLQDVC NRQCSDSTGY LEVSVRAAKQ 60 PQSELCFSVA NGSKFESKVD ERARDGERFR IERDQIRQYA QSIYRTLADP QAAAVPTEQR 120 SSGLRSSIPG GQLACDVSKY RVQTASANQL HPNSNDSFES INFSPSSPIT DHQRELRRTE 180 IEAGLEPEGT RRQNVGEEAS DQINRATSLK NGFPLAERAS APEKFIAEGL DDSEPARHGR 240 GGPQNKRSSI WAKNVELVGH GPFAFRENIV ECSTHIGVSE KEGFVLHNVI DAQASHGRAK 300 EPTADSGTPR SQNSHQRTIH PQGRQDRAKT ANAIPSSPQS QALTDRVSRG AASDALGTDL 360 LPQSIGSDLL QQGTTPARVE RVQNISRTRS GGERDVEDTL KGRRYPERIG DLRATTVATA 420 GRQHKAERSG EIPPVRTSQE TVVGTAGVLT IPERIGGTAE DLTAREIVDV SESHSVSERV 480 GGRAETHSAQ DGGSAEGFTI RERTGGSPSE GNKIPERIGA VSDRDSAGKV QEKGKEEKLH 540 PREEGHQRQS ESSDERRLRA RIDGSGGSAS GGGDEIEEPK VVRPARPSGG KDRHSKVNTA 600 KGPRDRRVRL SVPTAVQFYD VQDRLGFDQP SKAVEWLIHK AKDAIDELGR IPVDGKDGPR 660 VPSVCTSDIT RHVSISSSYG AGAVSGLGMF APLQPNQGNF AGPTFGVGRM DLTRPNSGSA 720 EGGRSNESAG SPLARVESRT KARERARERT KKGPGRDGNQ GKVTSGGSPS SSQSPPFHPN 780 PAFNSFNASN SIQSSGHGLS LQSPHQSALL PAAYDGSPFS RIMQGSHQAT FQNMIMHNPP 840 LDCQFGDQTQ SPLHNAFLVQ PQASLPSPSH TMNTLQHPHL QTMQVNAQGA LHLGTNFNPG 900 PCSSSFAEAS MSSPHYPPVP TLPISQSLSI PQLSSLGMSP ASSLGPHYPT GSPTPIPPGF 960 SPGPPASAFR AHSLSISQCS NPNQNHQQQQ QQLQGLSRVQ RPVPSFSTTP STSHAGSSSS 1020 RAHAHPYYSG NSSTALMGCL QSMPSFAPRS NVGYEHGQRV RPMMSPNRIH SYQASNSQIP 1080 ARIHGLDDME ELEEEPNPAT PSSPHLSPHP * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zkt_A | 7e-20 | 595 | 648 | 1 | 54 | Putative transcription factor PCF6 |
| 5zkt_B | 7e-20 | 595 | 648 | 1 | 54 | Putative transcription factor PCF6 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00203 | DAP | Transfer from AT1G53230 | Download |
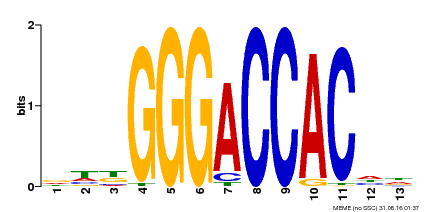
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A2R6XW14 | 0.0 | A0A2R6XW14_MARPO; Uncharacterized protein | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18390.2 | 3e-30 | TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0001s0298.3.p |



