 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0005s0212.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 372aa MW: 39140.1 Da PI: 6.1707 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 53.2 | 4e-17 | 269 | 303 | 1 | 35 |
GATA 1 CsnCgttkTplWRrgpdgnktLCnaCGlyyrkkgl 35
C +C t Tp+WR gp g+ktLCnaCG++y++ +l
Mapoly0005s0212.1.p 269 CMHCSTQRTPQWRAGPMGPKTLCNACGVRYKSGRL 303
99*****************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF016992 | 3.5E-70 | 9 | 356 | IPR016679 | Transcription factor, GATA, plant |
| SMART | SM00401 | 6.6E-14 | 263 | 313 | IPR000679 | Zinc finger, GATA-type |
| PROSITE profile | PS50114 | 11.213 | 263 | 299 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 1.85E-14 | 265 | 326 | No hit | No description |
| CDD | cd00202 | 1.26E-12 | 268 | 313 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 6.6E-14 | 268 | 301 | IPR013088 | Zinc finger, NHR/GATA-type |
| PROSITE pattern | PS00344 | 0 | 269 | 294 | IPR000679 | Zinc finger, GATA-type |
| Pfam | PF00320 | 6.2E-15 | 269 | 303 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009416 | Biological Process | response to light stimulus | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 372 aa Download sequence Send to blast |
MELSSTFAGA CPVIMGEAFH IDDLLDFSNE DIAGPIGEGL LSSLDSTITD VETISTTGSP 60 SLAGTKHILD GDDVVAGDLC VPCDDLAELE WLSKFVEDSF SMGEATRPVT SSGGLTVSTD 120 VQIESYTNKD RDRFQSASPV SVLESSASSE RASNLYRELS VPGRARSKRA RTGGRGWGTT 180 ARILSSSSNS GSGSGSGSGS GNDTESLSSD SNAMTGGGNS SSQTLSVDSQ ATPSAAMIAP 240 KAVSSSAKKP WKASKSGKKG QDASQPWRCM HCSTQRTPQW RAGPMGPKTL CNACGVRYKS 300 GRLLPEYRPA GSPGYIGHKH SNSHKKILEM RRQREQQQQS QPSSMLSSSS CQVSPNHLLE 360 LEDPDICNSQ S* |
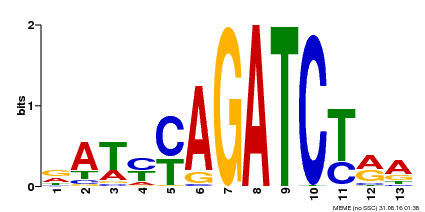
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00530 | DAP | Transfer from AT5G25830 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024358213.1 | 4e-89 | GATA transcription factor 9-like | ||||
| TrEMBL | A0A176VU14 | 0.0 | A0A176VU14_MARPO; GATA transcription factor | ||||
| TrEMBL | A0A1L7B539 | 0.0 | A0A1L7B539_MARPO; GATA transcription factor | ||||
| STRING | PP1S241_7V6.1 | 2e-88 | (Physcomitrella patens) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP68 | 17 | 287 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G32890.1 | 1e-34 | GATA transcription factor 9 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0005s0212.1.p |



