 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0006s0226.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 450aa MW: 48853.8 Da PI: 6.4459 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54 | 3.9e-17 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT eEd++l d++ ++G g+W+t ++ g+ R++k+c++rw +yl
Mapoly0006s0226.1.p 14 RGPWTREEDLKLTDYISKHGEGCWRTLPKNAGLLRCGKSCRLRWINYL 61
89******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 50.9 | 3.7e-16 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+ ++eEd l++ ++ +lG++ W++Ia +++ gRt++++k++w+++l
Mapoly0006s0226.1.p 67 RGNISEEEDNLIITLHSLLGNR-WSLIAGRLP-GRTDNEIKNYWNTHL 112
7899******************.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-23 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.459 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.49E-29 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.6E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.0E-14 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.25E-11 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 27.3 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.2E-26 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.7E-17 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-14 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.73E-12 | 71 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010224 | Biological Process | response to UV-B | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0090378 | Biological Process | seed trichome elongation | ||||
| GO:1903086 | Biological Process | negative regulation of sinapate ester biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 450 aa Download sequence Send to blast |
MGRAPCCEKV GLNRGPWTRE EDLKLTDYIS KHGEGCWRTL PKNAGLLRCG KSCRLRWINY 60 LRPDLKRGNI SEEEDNLIIT LHSLLGNRWS LIAGRLPGRT DNEIKNYWNT HLKKKLRSMG 120 IDPNSHRPLS QAAMAVGSHI SQESNVSPCC TLAPQQQIAA IKPNCKTFQS LSMAHQAELN 180 GAMMPQCIMK DESYQTMALV ESFLRDNTVY AESQALPQLR EHCSAATGGG CGGGGGGGGG 240 GAAATAPISH RSNYMSLDKS ESLSMLSPCS PVSILQHCSL STDREKSPSS QALTDFTTSH 300 SDFDHKQALG GMTFGESPSL TSESTASSGF LQVDRGTPTS YEPVPAAAAW GAQLSTSLPL 360 VNSLDMSADH RKRYQEATDI SSTNCPNFQK MLFNEDVLSP WDMEILQPLG DSLGDHAEMD 420 PNAFHCSSNN PATLWDSEMT HSNQLPTAF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 2e-29 | 12 | 116 | 5 | 108 | B-MYB |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 111 | 117 | LKKKLRS |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00053 | PBM | Transfer from AT1G22640 | Download |
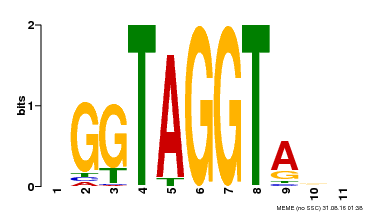
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A176VUA8 | 0.0 | A0A176VUA8_MARPO; Uncharacterized protein | ||||
| TrEMBL | A0A2R6XQ29 | 0.0 | A0A2R6XQ29_MARPO; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP5 | 17 | 1784 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G22640.1 | 6e-75 | myb domain protein 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0006s0226.1.p |



