 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0007s0056.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 466aa MW: 49937.1 Da PI: 7.5681 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 32.7 | 1.7e-10 | 295 | 353 | 5 | 63 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
kr++r NR +A rs +RK +i eLe+kv++L++e ++L +l +l+ l++e+
Mapoly0007s0056.1.p 295 KRAKRILANRQSAARSKERKMRYISELERKVQTLQTEATTLSAQLTMLQRDTTGLTTEN 353
9*********************************************9998887777766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 4.1E-17 | 291 | 355 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.151 | 293 | 356 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 2.95E-11 | 295 | 347 | No hit | No description |
| Pfam | PF00170 | 4.3E-9 | 295 | 352 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 4.1E-11 | 295 | 349 | No hit | No description |
| CDD | cd14703 | 9.82E-26 | 296 | 345 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 466 aa Download sequence Send to blast |
MDGLGDGYEA FVERLQSDFA ATSAAKNSGQ NGNTTTHSSH PPHPSQSQLH QSRYVGYPSP 60 THAFTVKREA SPSVSESSMR TANSLSIEVS MQDQVTSPGP PSVTGPSSSR NPSPANNYST 120 DVNQMPDTPP RRRGHRRAQS EIAFRLPDDV SFERDLGMHG SEMPALSDDA AEDLFSMYID 180 MEKINSFSAT SAPLPGPKST AADSGSTPPP THHSRSLSVD GVLSGFNSGR GSMGLSGGSA 240 GFPSDGRRSR HQHSSSMDGS TSFKHDLLAA ELDGADAKKV MNSAKLADIA LIDPKRAKRI 300 LANRQSAARS KERKMRYISE LERKVQTLQT EATTLSAQLT MLQRDTTGLT TENSELKLRL 360 QAMEQQAQLR DALNEALRDE VQRLKVATGQ MNGLTGHVYG LGQGPLTAQQ LQQLQQQSSQ 420 QQAHNDFLQR GTFGGLQTMS GQIGGSFIKS EGSTISVNQG SSASF* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor with an activatory role. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00126 | DAP | Transfer from AT1G06070 | Download |
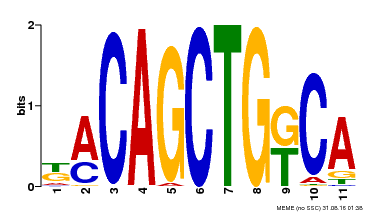
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024356505.1 | 0.0 | transcription factor RF2a-like | ||||
| Refseq | XP_024356506.1 | 0.0 | transcription factor RF2a-like | ||||
| Refseq | XP_024404099.1 | 0.0 | transcription factor RF2a-like | ||||
| Swissprot | Q04088 | 2e-87 | POF21_ARATH; Probable transcription factor PosF21 | ||||
| TrEMBL | A0A2R6XND5 | 0.0 | A0A2R6XND5_MARPO; Uncharacterized protein | ||||
| STRING | PP1S20_116V6.1 | 0.0 | (Physcomitrella patens) | ||||
| STRING | PP1S20_118V6.1 | 0.0 | (Physcomitrella patens) | ||||
| STRING | PP1S20_119V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP126 | 17 | 181 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06070.1 | 2e-83 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0007s0056.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



