 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0015s0005.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 436aa MW: 44816.1 Da PI: 6.9578 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 73.7 | 2.6e-23 | 297 | 359 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e+e+kr+rrkq+NRe+ArrsR+RK+ae+eeL ++v++L+ eN+aL+ el + ++e++k+k+++
Mapoly0015s0005.1.p 297 EREVKRQRRKQSNRESARRSRLRKQAECEELGTRVDTLTVENMALRTELARVSEECKKMKADN 359
799*********************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 3.6E-29 | 1 | 97 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 1.1E-7 | 141 | 193 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 1.3E-19 | 290 | 356 | No hit | No description |
| Pfam | PF00170 | 2.7E-21 | 297 | 359 | IPR004827 | Basic-leucine zipper domain |
| SMART | SM00338 | 2.7E-22 | 297 | 361 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 12.403 | 299 | 362 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.38E-10 | 300 | 356 | No hit | No description |
| CDD | cd14702 | 7.71E-25 | 302 | 352 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 304 | 319 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 436 aa Download sequence Send to blast |
MGTGETGTPT KAAKVSTTQE QQTTPTPYAE WGAAFQAYYN SGGQPPPGYF PPSVGSGPQP 60 HPYMWGGQPM MPGYGTPAPP YGAMYPHGGM YSHPSMPPGA HPYSQYGMPS PGGAIASEAT 120 ATTPAGGEGE GTKVADRLKN RLPLKRSKGS DGGKGNSGSA DGVFSQSGEG ESGSEGSSEG 180 SDDDNSQSML RQRSFEQMTL ESAIVVAGAP APAYSNQQPG FVNLAVGSGG AAGAQFGMPG 240 KGSGGSGTNL AGMNSGKKGK RPSASSSGGM VPTTPGTPLK GSSGRDGVPP ELWLQDEREV 300 KRQRRKQSNR ESARRSRLRK QAECEELGTR VDTLTVENMA LRTELARVSE ECKKMKADNA 360 ALFAQLRKQA NTNNVAEEAA EDAQAVQTEG NGKAQDGSPR RNPSRNEEEA SGDSPTKAPA 420 SAEAGTTPAE AVAAG* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 313 | 319 | RRSRLRK |
| 2 | 313 | 320 | RRSRLRKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00291 | DAP | Transfer from AT2G35530 | Download |
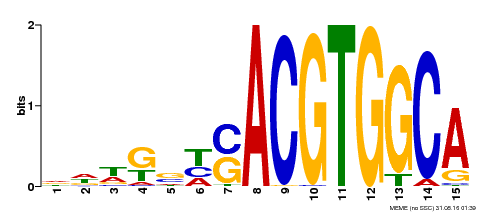
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010261558.1 | 6e-83 | PREDICTED: bZIP transcription factor 16 isoform X2 | ||||
| TrEMBL | A0A176VMX5 | 0.0 | A0A176VMX5_MARPO; Uncharacterized protein | ||||
| TrEMBL | A0A2R6XGE1 | 0.0 | A0A2R6XGE1_MARPO; Uncharacterized protein | ||||
| STRING | XP_010261549.1 | 5e-82 | (Nelumbo nucifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP700 | 17 | 67 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46270.1 | 5e-29 | G-box binding factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0015s0005.1.p |



