 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0026s0039.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 504aa MW: 55387.9 Da PI: 7.525 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 33.2 | 1.2e-10 | 200 | 242 | 4 | 46 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLk 46
+k+ rr+++NReAAr+sR+RKka++++Le +L + ++L+
Mapoly0026s0039.1.p 200 SKALRRLAQNREAARKSRLRKKAYVQQLESSRIKLNQLEQELQ 242
7999*************************86655555554444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.1E-4 | 197 | 250 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 8.8E-8 | 197 | 250 | No hit | No description |
| PROSITE profile | PS50217 | 9.473 | 199 | 248 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 6.7E-8 | 201 | 247 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14708 | 2.17E-21 | 201 | 247 | No hit | No description |
| SuperFamily | SSF57959 | 4.31E-7 | 201 | 249 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 204 | 219 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 5.1E-34 | 303 | 378 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009410 | Biological Process | response to xenobiotic stimulus | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 504 aa Download sequence Send to blast |
MANSRGQRGP GVPAPIASAS PPVLHGMQGL SSSPPHLIAS FGAEPIKDHT GGYDLGELEQ 60 PLSFNLDEEL DLGERHSTVT SRPTLHILQS GPFRFSDTAN KQRQQPNQQQ QQQQSSQVTS 120 PSAASTDSRS AHFTPRRHPS FESPSSRQFE HWGDSRMADN SPRTDTSTDM EADVKLEDAH 180 HGANAGSNAS DHEATKNGDS KALRRLAQNR EAARKSRLRK KAYVQQLESS RIKLNQLEQE 240 LQRARQQLMT VRGERVQGVY HLGGYGDQNP HGVGGNGVNG AINPGAAAFD MEYGRWVDEQ 300 IRQMCDLRAA LQAHVADNEL RVLVDGGMAH YDEIFRLKSI AAKADVFHLV SGMWKTPAER 360 CFMWMGGFRP SELLKILIPQ IEPLTEQQLL GICNLQQSSQ QAEDALSQGM EALQQSLADT 420 LAAGSLGSSP NVANYMGQMA MAMGKLGTLE SFVRQADNLR QQTLQQMYRI LTTRQAARGL 480 LAMGDYFARL RALSSLWSAR PRD* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
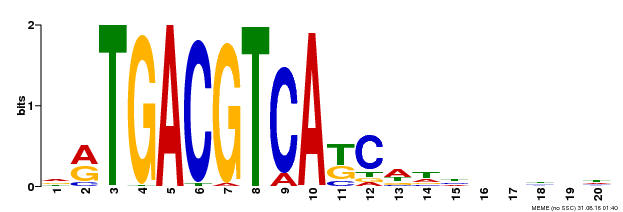
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-TGACG-3'. Recognizes ocs elements like the as-1 motif of the cauliflower mosaic virus 35S promoter. Binding to the as-1-like cis elements mediate auxin- and salicylic acid-inducible transcription. Binds to the hexamer motif 5'-ACGTCA-3' of histone gene promoters. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00492 | DAP | Transfer from AT5G06950 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024529114.1 | 0.0 | transcription factor TGA2.3 | ||||
| Refseq | XP_024536795.1 | 0.0 | transcription factor TGA2.3 isoform X1 | ||||
| Swissprot | Q41558 | 1e-156 | HBP1C_WHEAT; Transcription factor HBP-1b(c1) (Fragment) | ||||
| TrEMBL | A0A2R6XAM1 | 0.0 | A0A2R6XAM1_MARPO; Uncharacterized protein | ||||
| STRING | PP1S258_7V6.1 | 1e-180 | (Physcomitrella patens) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP722 | 16 | 67 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G12250.2 | 1e-131 | TGACG motif-binding factor 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0026s0039.1.p |



