 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0026s0039.4.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 400aa MW: 44335.6 Da PI: 8.474 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 32.5 | 1.9e-10 | 106 | 144 | 4 | 42 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeN 42
+k+ rr+++NReAAr+sR+RKka++++Le +L +
Mapoly0026s0039.4.p 106 SKALRRLAQNREAARKSRLRKKAYVQQLESSRIKLNQLE 144
7999************************98555444444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 1.1E-7 | 103 | 149 | No hit | No description |
| SMART | SM00338 | 8.1E-8 | 103 | 167 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.174 | 105 | 149 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 3.1E-7 | 107 | 146 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 4.98E-7 | 107 | 150 | No hit | No description |
| CDD | cd14708 | 8.67E-22 | 107 | 156 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 110 | 125 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 3.6E-34 | 199 | 274 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009410 | Biological Process | response to xenobiotic stimulus | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 400 aa Download sequence Send to blast |
MLTGMHQQRQ QPNQQQQQQQ SSQVTSPSAA STDSRSAHFT PRRHPSFESP SSRQFEHWGD 60 SRMADNSPRT DTSTDMEADV KLEDAHHGAN AGSNASDHEA TKNGDSKALR RLAQNREAAR 120 KSRLRKKAYV QQLESSRIKL NQLEQELQRA RQQGVYHLGG YGDQNPHGVG GNGVNGAINP 180 GAAAFDMEYG RWVDEQIRQM CDLRAALQAH VADNELRVLV DGGMAHYDEI FRLKSIAAKA 240 DVFHLVSGMW KTPAERCFMW MGGFRPSELL KILIPQIEPL TEQQLLGICN LQQSSQQAED 300 ALSQGMEALQ QSLADTLAAG SLGSSPNVAN YMGQMAMAMG KLGTLESFVR QADNLRQQTL 360 QQMYRILTTR QAARGLLAMG DYFARLRALS SLWSARPRD* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
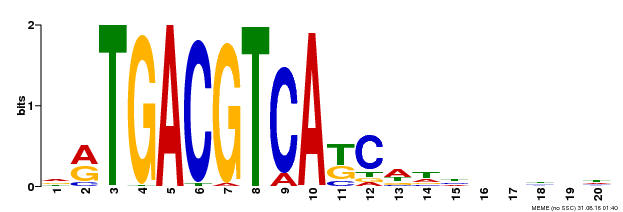
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-TGACG-3'. Recognizes ocs elements like the as-1 motif of the cauliflower mosaic virus 35S promoter. Binding to the as-1-like cis elements mediate auxin- and salicylic acid-inducible transcription. Binds to the hexamer motif 5'-ACGTCA-3' of histone gene promoters. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00492 | DAP | Transfer from AT5G06950 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024529114.1 | 0.0 | transcription factor TGA2.3 | ||||
| Refseq | XP_024536795.1 | 0.0 | transcription factor TGA2.3 isoform X1 | ||||
| Swissprot | Q41558 | 1e-161 | HBP1C_WHEAT; Transcription factor HBP-1b(c1) (Fragment) | ||||
| TrEMBL | A0A2R6XAL5 | 0.0 | A0A2R6XAL5_MARPO; Uncharacterized protein | ||||
| STRING | PP1S258_7V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G12250.2 | 1e-135 | TGACG motif-binding factor 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0026s0039.4.p |



