 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0032s0153.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 472aa MW: 50800.4 Da PI: 8.5618 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.1 | 3.7e-17 | 22 | 67 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+g+W++eEd l ++v ++G ++W++I++ ++ gR++k+c++rw +
Mapoly0032s0153.1.p 22 KGPWSPEEDAALQRLVDKYGARNWSLISKGIP-GRSGKSCRLRWCNQ 67
79******************************.***********985 PP
| |||||||
| 2 | Myb_DNA-binding | 52.7 | 9.6e-17 | 76 | 118 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
++T+ Ed +++a++ +G++ W+tIar ++ gRt++ +k++w++
Mapoly0032s0153.1.p 76 PFTAAEDAAIIQAHAHHGNK-WATIARLLP-GRTDNAIKNHWNST 118
89******************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 26.636 | 17 | 72 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.86E-31 | 20 | 115 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.1E-16 | 21 | 70 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.0E-17 | 22 | 67 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-24 | 23 | 75 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 5.71E-15 | 24 | 66 | No hit | No description |
| PROSITE profile | PS51294 | 21.649 | 73 | 123 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.1E-14 | 73 | 121 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 9.54E-12 | 76 | 119 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 4.9E-23 | 76 | 122 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 4.6E-14 | 76 | 118 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 472 aa Download sequence Send to blast |
MAVSPSTSLD FRCRTQDNER IKGPWSPEED AALQRLVDKY GARNWSLISK GIPGRSGKSC 60 RLRWCNQLSP QVQHRPFTAA EDAAIIQAHA HHGNKWATIA RLLPGRTDNA IKNHWNSTLR 120 RRYLAERSRA DEEGSYVCRI RDEDDLTSTL EARKQRCSID LESSGMAQQL SNEGSSFLDS 180 SSMCGSPLNT SQCAQQVKKL NFAPSSPSCS EHSDPTTVLA PQVFRPVPRP SAFTCFSPSS 240 TTSGGLTKQM QQSPQEASSS STDPPTSLSL SLPGTCANRI EREVSPRCSP TTRPTLPPPQ 300 PIAQAQHSPR SENHHHLRPH EQLKESPPHI SAIAWRQPAP PAEHLHEQSG FGIVNNGGAM 360 LVGQAAVPTD MMSAAVRAAV AQALQAPAAA APPARAAPMM GMGFGLDAAV NAGLLAMMRD 420 MVAKEVQKYM AAVQAPTCLP PFSVMGPDYA SLTSHPEFLG TMTPSVPRKA G* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 4e-39 | 19 | 122 | 1 | 104 | MYB PROTO-ONCOGENE PROTEIN |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00400 | DAP | Transfer from AT3G50060 | Download |
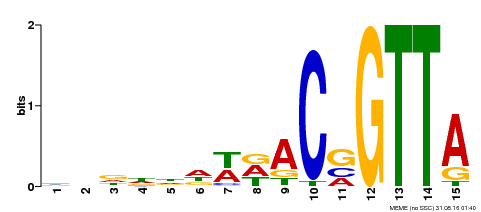
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024362152.1 | 1e-105 | transcription factor MYB77-like | ||||
| TrEMBL | A0A2R6X7C2 | 0.0 | A0A2R6X7C2_MARPO; Uncharacterized protein | ||||
| STRING | PP1S10_267V6.1 | 1e-104 | (Physcomitrella patens) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP5 | 17 | 1784 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G67300.1 | 6e-52 | myb domain protein r1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0032s0153.1.p |



