 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0036s0107.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 1109aa MW: 118670 Da PI: 6.5384 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.1 | 7.2e-17 | 56 | 102 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEde+l +av+ + g++Wk+Ia+ ++ Rt+ qc +rwqk+l
Mapoly0036s0107.1.p 56 KGGWTPEEDEILRRAVQCFKGKNWKKIAEFFQ-DRTDVQCLHRWQKVL 102
688*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 56.6 | 6.1e-18 | 108 | 154 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd+++ ++v ++G + W+ Ia+ ++ gR +kqc++rw+++l
Mapoly0036s0107.1.p 108 KGPWTKEEDDRIMELVGKYGAKKWSVIAQNLP-GRIGKQCRERWHNHL 154
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 57.7 | 2.8e-18 | 160 | 204 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r++WT++Ed+ l++a++ +G++ W+ Ia+ ++ gRt++++k++w++
Mapoly0036s0107.1.p 160 RDAWTQQEDLALIHAHQIYGNK-WAEIAKFLP-GRTDNSIKNHWNST 204
789*******************.*********.***********976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.226 | 51 | 102 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.28E-17 | 54 | 112 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.7E-14 | 55 | 104 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.8E-15 | 56 | 102 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.6E-23 | 58 | 118 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 5.00E-13 | 59 | 102 | No hit | No description |
| PROSITE profile | PS51294 | 32.302 | 103 | 158 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.95E-32 | 105 | 201 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.3E-16 | 107 | 156 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.1E-17 | 108 | 154 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.17E-14 | 110 | 154 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-26 | 119 | 161 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.9E-18 | 159 | 207 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 23.754 | 159 | 209 | IPR017930 | Myb domain |
| Pfam | PF00249 | 2.6E-16 | 160 | 203 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.6E-24 | 162 | 209 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.84E-13 | 162 | 202 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1109 aa Download sequence Send to blast |
MFMIRAGGAM ATQDSQNKMR TGPVPSSSGA ESEDDAIHQL PPVHGRTGGP TRRSSKGGWT 60 PEEDEILRRA VQCFKGKNWK KIAEFFQDRT DVQCLHRWQK VLNPDLVKGP WTKEEDDRIM 120 ELVGKYGAKK WSVIAQNLPG RIGKQCRERW HNHLNPNIKR DAWTQQEDLA LIHAHQIYGN 180 KWAEIAKFLP GRTDNSIKNH WNSTMKKKVD PMSATDPISR ALAAYQAQQD SLKSGGSANS 240 EQSMCLSNTV DVVLPSTSGV SNSNLSARIM ASSTVPPSVR TCDVAEDSAV DSKPAIQLAA 300 PAGSQVPEKD DIKAVKSGSR KELGTASKPP LLPPPPPPRA SCAGNPLTQS TCDTVNCTPM 360 AVPVVSTSSS SLQRQPSSID ENVCVPSSVE LCRVPSFSRM YPLACSLPES TTCSGQSTPA 420 LTSLPLTSSV FGNGTMIGDV DSVSGSVIES VNMMAQPMTP MGYMTLHECH YSQHQRAEEL 480 SGPCLPPYSN NQKLCLRGDI SKVGTEEYSD ELLKAATQVV SDYQPLSMFA ELEPEVEDTE 540 GDAGFFYEPP RLPNSDMPFL NYDLISPNGT SLQAYSPLGV RQMIMPSVNC STPPSYPSSS 600 PFRGGSPQST LRRAAQSFGG TPSILRKRPR PLVSISRGNK EGEASGLQVR DSNMSGGCVQ 660 SSDNAADMAH DSACTVTPDR NTSNENSFLC RSFGSDSLMS ADISPCHARR FASPAYDLTS 720 KNLMKINRLE IENDFQSGSG YPSPGSNSSG GAHTRTKGGD GKSGGCVSGI KCNKRGRFRA 780 INPDYESKAI KGGGNQPSPK TQDLPVLVEQ RQNGQQACFS VEASSMTSCV LMAPGAEAYG 840 GAGSFGGVTG RYGCCDSKSQ YEAGHVGSHA AVVGSQPKGS DKRENMALNS VSSFNAWGSA 900 ISPTVSGSNH GGKDWFNALA DFDTFNFALS GDGNSPSAIW RSPWKLDPAV PKDGVMSLLE 960 GMDMLVEDGS DDALGIMKHI SEHAAPAYSE AQEVLAKSVL MECHGRAASP NPVHRSRQEG 1020 SNEYKENVRS MNEHVDNCPS SPFVSWIVPF PAEMLSPDAA AGGTPLRGSS LGHSLQVAGG 1080 VDDNQLTFGG VSMVDPFSPS LYLLRECR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 1e-72 | 56 | 209 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-72 | 56 | 209 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 626 | 630 | KRPRP |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
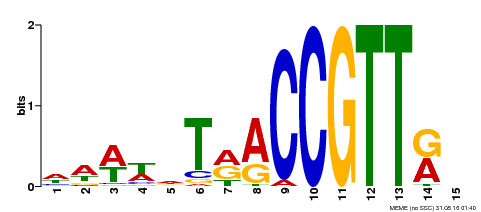
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF190310 | 1e-63 | AF190310.1 Marchantia polymorpha c-myb-like transcription factor (MYB3R-1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A2R6X4U0 | 0.0 | A0A2R6X4U0_MARPO; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP5 | 17 | 1784 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G11510.2 | 1e-90 | myb domain protein 3r-4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0036s0107.1.p |



