 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0069s0009.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 378aa MW: 41496.2 Da PI: 9.6524 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 48.5 | 1.9e-15 | 301 | 350 | 5 | 54 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkk 54
+r+rr++kNRe+A rsR+RK+a++ eLe +v +L++eN +L+ke+ee +k
Mapoly0069s0009.1.p 301 RRQRRMIKNRESAARSRARKQAYTVELEAEVSQLKEENNRLRKEMEEASK 350
79******************************************998766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 8.0E-15 | 290 | 353 | No hit | No description |
| SMART | SM00338 | 3.1E-13 | 297 | 358 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 12.001 | 299 | 351 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.3E-13 | 301 | 351 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14707 | 1.38E-23 | 301 | 355 | No hit | No description |
| SuperFamily | SSF57959 | 3.38E-11 | 301 | 350 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 304 | 319 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0010152 | Biological Process | pollen maturation | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 378 aa Download sequence Send to blast |
MMASTSGGAQ MGTRNVPFTP ITRQNSIFSL TLDELQNAID PGKHFGSMNM DEFLKNVWSA 60 EENQTMSLAI STSNESNAPQ SSSATGGGLA RQGSLTHPGS LSLSRTLSRK TVDEVWRDIH 120 RVPEPTAKEQ QRANNYGEMT LEDFLVKAGV VREDSESGSP ASNNTNTFTA PFAAPLASQN 180 PFHGPTVLAQ STHQAAMEAA IAHNQHQQHV EWHNYQLKSN AMQHQQHVLQ QQAQEAAAAY 240 AMPKRGPVIA LPGTPSGSGL EGSSDGGLQV NGLSPAITTP DTPNRGRKRG PESMPEKTVE 300 RRQRRMIKNR ESAARSRARK QAYTVELEAE VSQLKEENNR LRKEMEEASK RKREELLIQE 360 VPPRKNQALR RSCSISW* |
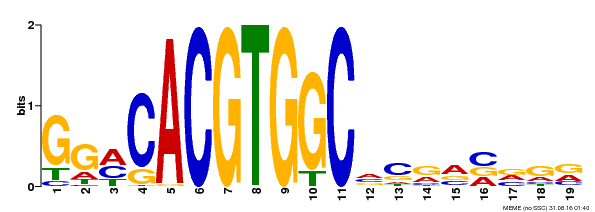
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00087 | SELEX | Transfer from AT1G49720 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A176W1D2 | 0.0 | A0A176W1D2_MARPO; Uncharacterized protein | ||||
| TrEMBL | A0A2R6WP76 | 0.0 | A0A2R6WP76_MARPO; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP540 | 15 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49720.2 | 6e-32 | abscisic acid responsive element-binding factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0069s0009.1.p |



