 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0072s0031.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 600aa MW: 65419.3 Da PI: 5.0545 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 151.7 | 5.7e-47 | 43 | 165 | 2 | 124 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspe 91
+ r+p+ +ErEnnk+RERrRRa+aa+i+aGLR++Gny+lpk+aD neVlkALc+eAGw+ve+DGt yr+g k +e ++ ++s+ ++++
Mapoly0072s0031.2.p 43 SVLRMPSPRERENNKKRERRRRAVAARIFAGLRQYGNYRLPKHADHNEVLKALCAEAGWIVEEDGTIYRQGHKTPELKADLQESEVINVN 132
5679***********************************************************************888888888886665 PP
DUF822 92 sslqsslkssalaspvesysaspksssfpspss 124
+ + + p ++ + + s+ps+s
Mapoly0072s0031.2.p 133 CTCEIQEAALDSDGPESALDRFKYETSTPSSSC 165
554433333333334444444444444444433 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 6.7E-44 | 46 | 175 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001046 | Molecular Function | core promoter sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 600 aa Download sequence Send to blast |
MANRAAEASS SYNFKVRGCI KSTSGPWIVR RPPTKNSGRG ATSVLRMPSP RERENNKKRE 60 RRRRAVAARI FAGLRQYGNY RLPKHADHNE VLKALCAEAG WIVEEDGTIY RQGHKTPELK 120 ADLQESEVIN VNCTCEIQEA ALDSDGPESA LDRFKYETST PSSSCGQSDE TTCSIRSRNS 180 GRNNCDQPTV AGICDYDLNL SGTEGDYEEL DQVAVENEES TVMMSGLPEP KNLETSAEDC 240 DTNIEFFINP ESRPMNLPKS ERSTGECNAS YHQEFQFFAG LFPGPSDLLT AALAVADDTP 300 SCCVRPTRDL NSPPIFEGST STEFPNAVNC SLPPAPSMDN TFIKKESYND QIYLKNMGRQ 360 DSTWIPACND HNGRSGDADS SPSSSDTPEL ELLSSGDVMI DSTSPCRMDA GRVSCSDASD 420 CPCAISDETP LQSSVSNGLL IARNYRVPPV SYAGPDQGYL LDRKNGSSSA LSNQRSMQST 480 REYSNWNDLT HRASGRGGQT AAATNIGHHQ SVSITTAASK PFDKTALYEF SPGTDFHGQN 540 KQQYRGPLIL PPKQQQLQSA PPVDALSLTL CTSTGSTGLS SEYPFCFIKR HIKLPIPQT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 2e-16 | 46 | 115 | 372 | 441 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 2e-16 | 46 | 115 | 372 | 441 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 2e-16 | 46 | 115 | 372 | 441 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 2e-16 | 46 | 115 | 372 | 441 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00073 | ChIP-chip | Transfer from AT1G19350 | Download |
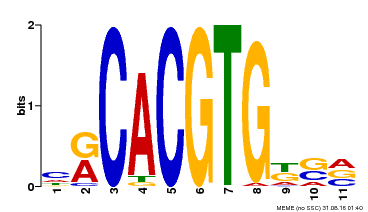
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A176VHR6 | 0.0 | A0A176VHR6_MARPO; Uncharacterized protein | ||||
| TrEMBL | A0A2R6WN42 | 0.0 | A0A2R6WN42_MARPO; Uncharacterized protein | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19350.6 | 8e-22 | BES1 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0072s0031.2.p |



