 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0072s0102.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 1165aa MW: 126793 Da PI: 8.0885 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 38.5 | 3e-12 | 366 | 414 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV + +grW A+I++ ++r++lg+f +e+Aa+a+++a+ k++g
Mapoly0072s0102.1.p 366 SQYRGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNREEDAARAYDRAAMKFRG 414
78****9888.8*********3.....5************************998 PP
| |||||||
| 2 | B3 | 97.9 | 6.1e-31 | 501 | 601 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.....---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE- CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.....ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkld 85
f+k+ tpsdv+k++rlv+pk++ae++ ++ +++ tl++ed sg+ W+++++y+++s++yv+tkGW++Fvk+++L++gD+v F+
Mapoly0072s0102.1.p 501 FEKAVTPSDVGKLNRLVIPKQHAERCfpldlALNAPCQ-TLSFEDVSGKHWRFRYSYWNSSQSYVFTKGWSRFVKEKKLEAGDTVSFE-- 587
89****************************98888885.*************************************************.. PP
SSSEE..EEEEE-S CS
B3 86 grsefelvvkvfrk 99
+ ++el++ ++r+
Mapoly0072s0102.1.p 588 RGPNQELYIDFRRR 601
44889999999986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 3.08E-14 | 366 | 424 | No hit | No description |
| SuperFamily | SSF54171 | 3.07E-17 | 366 | 422 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.6E-7 | 367 | 414 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 5.4E-29 | 367 | 428 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 20.455 | 367 | 422 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.6E-21 | 367 | 422 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 4.9E-39 | 496 | 600 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 8.5E-30 | 498 | 597 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 2.60E-27 | 499 | 600 | No hit | No description |
| Pfam | PF02362 | 1.6E-27 | 501 | 601 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 15.016 | 501 | 602 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 9.2E-26 | 501 | 602 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1165 aa Download sequence Send to blast |
MMTAATSRVT CLRLHSDPKE IADLWSNAVL RSQRSPLSTP GKGGPDVTLR LRAVWDQQSS 60 TSNTTDASLQ QQMTSPRLNR HGLTLNHEAA PGARTTQSST RSCDNSSSSS PPEQQQQQQQ 120 QHQQQQHHHH EHMKSLNASI NINTAVNSSS SDDKELVTVE RKTAKLEIGE HHHHLHHLHN 180 LHHHHFHQQQ MVKSSQPIKV EWAERGAAAA AAAGAGSGIN LANFSQSHKL EVSGDVKDEG 240 AASEECSSSP CSSSDKRMNL ELGACSQDDA AAAALVGVAA AAAASAAAGE RGGERGPPSW 300 AGTAWGPGCV TNPRSSSCLN ERAASERSFS GVSGDAVTGP PEESSDGGED SRGGVLKEGV 360 NKLPSSQYRG VVPQPNGRWG AQIYEKHQRV WLGTFNREED AARAYDRAAM KFRGRDAMTN 420 FRPVQETDPE AVFLRQYSKE QIVEMLRRHT YEEELEQSKR LASMRVTSPQ TPAGMVPNPP 480 NESDDPPSLP PKPTMQREHL FEKAVTPSDV GKLNRLVIPK QHAERCFPLD LALNAPCQTL 540 SFEDVSGKHW RFRYSYWNSS QSYVFTKGWS RFVKEKKLEA GDTVSFERGP NQELYIDFRR 600 RLNNQVAQML PGPSTSASDF ARNRPWVPRL PNPAGGLNPA VWHQLSLQNL QSSLESREQL 660 QMLYARQAFP QHMHSQQLSS FSSLQQQQQQ QQRQQQQHME SGVSLPHQTR TGDDFFSLLE 720 LGRRTDGGVA AMDRTVVGAS TTVVKPQQLL AEGPVSTPSA APAGASGGTR LFGVDLEKTS 780 SSSAQRSTER ESAVVSGLND WKSQTYAQQL QSLSRASSYH QGESSFQQQQ QQHLPTARIL 840 PSFQGSLHGQ SPAPVLMAAS AKQQQQQQQE QSSFMGLSIV RGRAGEQSPG TRLAASHSNL 900 QLASSTNMNS GSAARAAGAG AELQEAGART VVALGLAGPQ QFAGDSRKRK VGILESLKNS 960 GPADSYARDG APDTSLSLAQ HSESTTQVSF TTESSHFSSS TQPSGSTSTA EDREQKSPKR 1020 HCSPVQGAPT GSSKACAAGL PLTSTTTDTR GCRVDQKQQQ QQPPQPVASA NTTSGPPGAV 1080 VKCHLQWSPE KFLNISLANF HSVEGLKREL LQFTHLDFAQ GLDIVYKDKD GDIMLLSEHS 1140 WGLFKENVQE MWIRKTEQPQ QQRA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5os9_A | 3e-46 | 497 | 601 | 4 | 113 | B3 domain-containing transcription factor NGA1 |
| 5os9_B | 3e-46 | 497 | 601 | 4 | 113 | B3 domain-containing transcription factor NGA1 |
| Search in ModeBase | ||||||
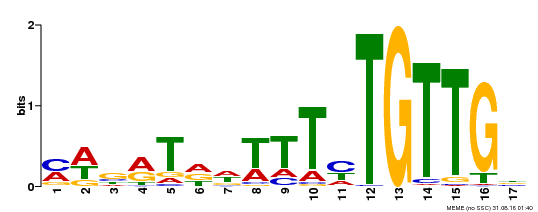
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00139 | DAP | Transfer from AT1G13260 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF542555 | 0.0 | AF542555.1 Marchantia polymorpha PAC pMM2D3 RS/SP6 region hypothetical alliin lyase-like protein (M2D3.1), hypothetical protein (M2D3.2), M2D3.3 protein (M2D3.3), and hypothetical protein (M2D3.4) genes, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A2R6WND4 | 0.0 | A0A2R6WND4_MARPO; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP365 | 16 | 108 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13260.1 | 5e-83 | related to ABI3/VP1 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0072s0102.1.p |



