 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0092s0058.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 415aa MW: 42421.5 Da PI: 4.8632 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 59.4 | 8.4e-19 | 217 | 266 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
+++GVr+++ +g+W+AeIrdp + r++lg+f t+e+Aa+a++ a+++++g
Mapoly0092s0058.1.p 217 HFRGVRRRP-WGKWAAEIRDPE----KgVRVWLGTFPTPEKAARAYDCAARRIRG 266
69*******.**********73....24*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.730.10 | 1.6E-31 | 216 | 276 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.11E-21 | 217 | 276 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 6.94E-30 | 217 | 275 | No hit | No description |
| PROSITE profile | PS51032 | 24.184 | 217 | 274 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 9.4E-13 | 217 | 266 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.7E-36 | 217 | 280 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.7E-10 | 218 | 229 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.7E-10 | 240 | 256 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0034059 | Biological Process | response to anoxia | ||||
| GO:0071456 | Biological Process | cellular response to hypoxia | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 415 aa Download sequence Send to blast |
MTMVVADDGP VGGGDEWWHR KRSRSCFDDG GDDGAGAAAA AAGGAAAGGD DLGSARRART 60 PGSPDERFLA MRGGGAGGEE GGGGGGAGRA REEEEDDGAS VSCCCATSSS CGSTMRDAGA 120 DGAGGGEVLD PGVEKLDPKS SSLRDFGVSP ASGSASVAAA AIKDAAGKGP SLSSGSSCLS 180 SNSCASNKAT KVEAAAAPST AAAAGGAAPA AETTPRHFRG VRRRPWGKWA AEIRDPEKGV 240 RVWLGTFPTP EKAARAYDCA ARRIRGKKAK LNYPDENSPT KPLTKQVQKV VKKKPQSPVS 300 VVQGPLSSST ESNDSRVSPP ASREVAGEVD LASLDSGSVA DCLDSCSVGD GSTVTEVGGE 360 VEHQHAQYDT SHMEEFQIED KVRARDLWQW SSLEPLCFDY AIPLWTFDDL PNLS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 5e-20 | 216 | 274 | 4 | 63 | ATERF1 |
| 3gcc_A | 5e-20 | 216 | 274 | 4 | 63 | ATERF1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00230 | DAP | Transfer from AT1G72360 | Download |
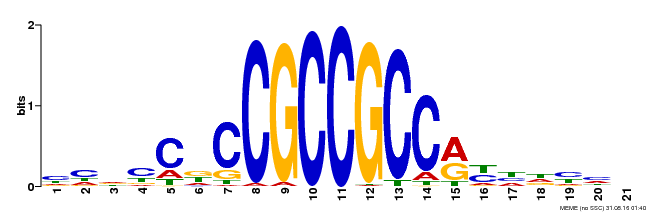
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A176WP64 | 0.0 | A0A176WP64_MARPO; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP6 | 16 | 1718 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72360.1 | 9e-28 | ERF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0092s0058.1.p |



