 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0119s0041.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 542aa MW: 59066.9 Da PI: 6.3636 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.8 | 8.8e-32 | 306 | 361 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k+r++W+peLH+rFv+a++qLGGs+ AtPk+i+elmkv+gLt+++vkSHLQkYRl+
Mapoly0119s0041.1.p 306 KARRCWSPELHRRFVSALQQLGGSQIATPKQIRELMKVDGLTNDEVKSHLQKYRLH 361
68****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.714 | 303 | 363 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.0E-29 | 303 | 364 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.51E-17 | 304 | 364 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.1E-26 | 306 | 361 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.2E-7 | 308 | 359 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009266 | Biological Process | response to temperature stimulus | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010452 | Biological Process | histone H3-K36 methylation | ||||
| GO:0048579 | Biological Process | negative regulation of long-day photoperiodism, flowering | ||||
| GO:1903507 | Biological Process | negative regulation of nucleic acid-templated transcription | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 542 aa Download sequence Send to blast |
MGSPADLTLG CRPQSSGNEF KAVSTSGDQI ERVRKLDEYL KALEDERHKI EAFKRELPHC 60 MQLLDYAIEV SKDRLVDSPR SSSGGGHAVD SYGEVETSTL GSFGKHVVEE FLKKSWEAPK 120 REIEEVEKVS KRSDEGRDAD HRPSWMAEAQ LWSQPSTRSD AWKEKESSTE DSTRRQRDTT 180 EQSTVTSPAN LFNTHKRTGG AFLPFIKEKP VATVASRVKV LSGADLALSS VESRPASRIA 240 LGPVESEAGS LDVSTTRTRD VGMEVVQAKD NGRKLNGSGS GGNHSGGTGS TSSGGGGGGS 300 GQAQRKARRC WSPELHRRFV SALQQLGGSQ IATPKQIREL MKVDGLTNDE VKSHLQKYRL 360 HTRRPSPAPQ APQQQAPQLV VLGGIWVPPE YTSGGMYHDP SSSVSQQGHF CQASLSQEIY 420 SQLNPSAQIQ LHRSQSHSSP QGPLQSTSQL SSGARQTSAE MYREDSPGED GKSESSSWKG 480 EDDSRGRSEK ESDREVGSGG RDDDEDDTDV EDEGRESGMR LKLEMAFTRS HTVMKPEPQM 540 A* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 4e-15 | 306 | 359 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 4e-15 | 306 | 359 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 4e-15 | 306 | 359 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 4e-15 | 306 | 359 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 4e-15 | 306 | 359 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 4e-15 | 306 | 359 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 4e-15 | 306 | 359 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 4e-15 | 306 | 359 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00261 | DAP | Transfer from AT2G03500 | Download |
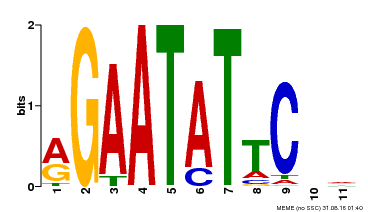
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A2R6WAI8 | 0.0 | A0A2R6WAI8_MARPO; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP4303 | 16 | 24 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G03500.1 | 3e-67 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0119s0041.1.p |



