 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0130s0030.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 855aa MW: 90841.8 Da PI: 5.4518 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 37.7 | 4.5e-12 | 284 | 343 | 2 | 61 |
XXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 2 kelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklks 61
+ kr+ r+++NRe+A+ sRqRKk +++eLe + ++ a +L ++ +l+ e +l+
Mapoly0130s0030.1.p 284 DDEKRQARLMRNRESAQLSRQRKKVYVDELEGRLRTMAATIAELTATITHLTAENVNLRR 343
567************************************************999888775 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 4.8E-14 | 283 | 347 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.117 | 285 | 345 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.7E-10 | 286 | 344 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.56E-11 | 287 | 345 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 9.8E-16 | 287 | 345 | No hit | No description |
| CDD | cd14704 | 3.12E-20 | 288 | 338 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006990 | Biological Process | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response | ||||
| GO:0009408 | Biological Process | response to heat | ||||
| GO:0005635 | Cellular Component | nuclear envelope | ||||
| GO:0005654 | Cellular Component | nucleoplasm | ||||
| GO:0005783 | Cellular Component | endoplasmic reticulum | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0048471 | Cellular Component | perinuclear region of cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 855 aa Download sequence Send to blast |
MEVEVVEADN GVNPNNDSSM AIDMAIPGGF EDENALMMDS SVETADLGED WVSLFFDESY 60 LNAMTTSGEV EQQFLPGSDA PAAAPGSIGN GIQISNRAVS PSTMILSSSP ASASSGCPFS 120 ANVDIYPENH NATDKTPLPT AAAISPPPHP AGSPESTLSQ ASGASSCAVQ PCWWGNEDLE 180 EDGVHGSEEE SSTSNYGATD PSAVAPDASK APIQAQEVKG GAEPASSVGK IATGARGSVE 240 RSDRKRRSWE REEAENGQQQ SQCAQSEIDR KGSESGDGLG GEEDDEKRQA RLMRNRESAQ 300 LSRQRKKVYV DELEGRLRTM AATIAELTAT ITHLTAENVN LRRQLGFFYQ PARPAGGMPI 360 MQYPGLVRPV YPGAPMPPVP IPRLKPQQSL TKTPKRAKST REPGEKKKVK KVASLATAGV 420 MGLLCITMLL GPFDWGSSTV SSGTTHIHVG NAQIRVGGRV LTSWNEEGVE DLRNMTPREP 480 WADGGGGNIG HVNSAGAGGR NGVGGRARSR IDRRNGNILR RNFSAGTERV WQSCSECYSR 540 RDEPRTAPNL VEKLVATNVS EQVSLIVPRN NRLIKVDGNL IIRAVMAGDK AAEDSGKEEH 600 SSQEGENGRG QVKQDLSRSS RSPPTPADHR LIKAISTQAQ EGGGIVVGER QSRNVGAIRE 660 DKNVQKPAYK SDALASVSAT PTFSQLVLGS LAGPVLSTGM CTEVFRFDTS PTRTAPNTET 720 SRGSSSVAED QVSGVREEKT SAKTTEEPST RKKRDPYAVP LPPIRRQDGN GTEPVVDKVF 780 KGQQSDTEGH DASTMVVSVF TGPQDVSAED SRTGGGPAKG LSQIFVVVLV DSVKYITYSC 840 MLPPTGSQVH VVAS* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00345 | DAP | Transfer from AT3G10800 | Download |
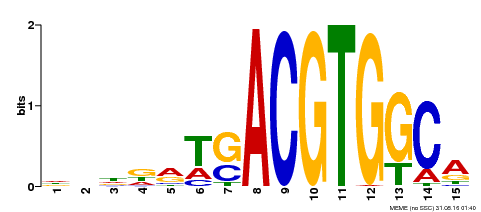
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A2R6W8A5 | 0.0 | A0A2R6W8A5_MARPO; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP2171 | 16 | 28 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40950.1 | 2e-24 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0130s0030.1.p |



