 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Mapoly0174s0004.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Marchantiophyta; Marchantiopsida; Marchantiidae; Marchantiales; Marchantiaceae; Marchantia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 829aa MW: 89587.6 Da PI: 6.6468 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 53.1 | 5.7e-17 | 372 | 421 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f+ Lrel+P++ ++K +Ka+ L +++eYI+ Lq
Mapoly0174s0004.1.p 372 RSKHSATEQRRRSKINDRFQMLRELVPHS----DQKRDKASFLLEVIEYIQVLQ 421
889*************************9....9*****************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 2.36E-17 | 367 | 429 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.634 | 370 | 420 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.3E-21 | 370 | 433 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.7E-14 | 372 | 421 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.12E-15 | 372 | 425 | No hit | No description |
| SMART | SM00353 | 2.4E-12 | 376 | 426 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 829 aa Download sequence Send to blast |
MAFSSSGTPM SMKEHSLGVS IGPGASVGVS SAGYTTFAPA SHDFQGPVEF SQQQREYLAQ 60 RSMMGQAAEQ QHSQGRGQGG GHSQGQHINM NEGAKPSTVS THDFLSLYAD NSNQYAEHHD 120 NMRGLSSALT TKDFLQPLET RGNHAAFREG QPSRGEKHPR QPEGLPHGST PAPPAVVERL 180 LSTGMNVYNG GHGVPNYGRV AKGAIDLPAN TMVRMVGEGN VEMRGRGGFN PSAGSMVTRE 240 DHDARNDLPK DQDAAPRGDN LAYWSNVRMK FGMLPPELNY HSQPNQPTSH VVMENVQQPP 300 VPNMPKPWPQ GVQGVIDMTK RPTRVLSEDD EDDEEEYGDR PRETRSKEPL SKADPAQKVD 360 GKAPENRGGT PRSKHSATEQ RRRSKINDRF QMLRELVPHS DQKRDKASFL LEVIEYIQVL 420 QDKVRKFETA EQGRHQDRLK NMVWDLCKRG GSGPGDTSIE ATCMDALAYA KEFVPSSAGL 480 PIDVKQVTEA AEVVRQRAVA NADENGSTEL SHNPQHTHKS GHHGASSSHD EKEGAAVLGK 540 SPLSSQQHQG QTTFSSRNLQ HASQSKQLSA ASDAQANVAG EGQVQNSSTI APATGAYDQH 600 DKANEVEPSQ QTRTQASTQQ SQGEGDTSDQ QHPDTRHRHG TTTAEDTGNG HAASFTDDQS 660 NTKPHMKSDR QHGYGSSPRS THNSQATDTS QANWQGQQHG SSAENSQQKE ESRGATTSAQ 720 GQDPLTIQGG VINVSSVYSQ GLLDTLTRAL QSSGLDLSQA NISVQIDLGK SISSAALPPS 780 TPKVSGQPAE TSGQRHQQQS ESHTRQQGGS STDPEHPPLK RPKLEMEL* |
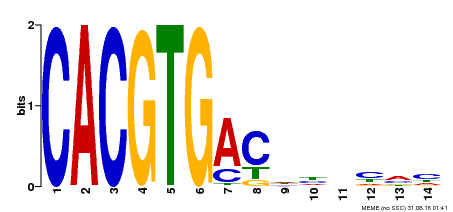
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024392458.1 | 2e-94 | uncharacterized protein LOC112290452 | ||||
| TrEMBL | A0A2R6W2K4 | 0.0 | A0A2R6W2K4_MARPO; Uncharacterized protein | ||||
| STRING | PP1S37_126V6.1 | 8e-94 | (Physcomitrella patens) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP1360 | 17 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.1 | 8e-30 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Mapoly0174s0004.1.p |



