 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr1g024025.1 | ||||||||
| Common Name | MTR_1g024025 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 1266aa MW: 141164 Da PI: 5.111 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 75.5 | 6.2e-24 | 146 | 247 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrs 88
f+k+lt sd++++g +++p++ ae+ +++++ ++l+++d++ ++W++++iyr++++r++lt+GW+ Fv +++L++gD+v+F +++
Medtr1g024025.1 146 FCKTLTASDTSTHGGFSVPRRAAEKLfppldyTIQPPT-QELVVRDLHDNTWTFRHIYRGQPKRHLLTTGWSLFVGSKRLRAGDSVLFI--RDE 236
99*********************999*****9555555.49************************************************..457 PP
EE..EEEEE-S CS
B3 89 efelvvkvfrk 99
+ +l+v+v+r+
Medtr1g024025.1 237 KSQLRVGVRRA 247
7778****997 PP
| |||||||
| 2 | Auxin_resp | 112.8 | 3.2e-37 | 272 | 355 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+++s+F+++YnPra++s+Fv++++k++k+++ +++s+GmRf m+fete+s +rr++Gt+vg+sd+dp+rWp+SkWr+++
Medtr1g024025.1 272 AAHAAANRSPFTIFYNPRACPSDFVIPLAKYRKSVYgTQLSAGMRFGMMFETEESGKRRYMGTIVGISDVDPLRWPGSKWRNIQ 355
79**********************************9********************************************986 PP
| |||||||
| 3 | Auxin_resp | 36.3 | 2.5e-13 | 898 | 940 | 1 | 45 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRf 45
aahaa+++ +F+++YnPra+ s+Fv++++k + + ++svGmRf
Medtr1g024025.1 898 AAHAAANRIPFTIFYNPRACLSDFVIPLAKSVYGT--QLSVGMRF 940
79**************************9877665..9******9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 2.62E-46 | 136 | 275 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.4E-41 | 138 | 260 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 9.66E-23 | 145 | 246 | No hit | No description |
| SMART | SM01019 | 6.3E-25 | 146 | 248 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.056 | 146 | 248 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 2.1E-21 | 146 | 247 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 5.0E-32 | 272 | 355 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 5.4E-11 | 743 | 841 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 23.746 | 754 | 838 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 6.57E-8 | 755 | 833 | No hit | No description |
| Pfam | PF06507 | 1.1E-7 | 898 | 940 | IPR010525 | Auxin response factor |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0010305 | Biological Process | leaf vascular tissue pattern formation | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048507 | Biological Process | meristem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1266 aa Download sequence Send to blast |
MASLDEKAKS VGGVVGGQTL VAEMKLLKEM QEHCGVRKTL NSELWHACAG PLVSLPQVGG 60 LVYYFPQGHS EQVAASTRRI ATSQIPNYPS LPSQLLCQVQ NVTLHADKET DEIYAQMTLQ 120 PLNSEKEVFP VSEFGLSKSK HPTEFFCKTL TASDTSTHGG FSVPRRAAEK LFPPLDYTIQ 180 PPTQELVVRD LHDNTWTFRH IYRGQPKRHL LTTGWSLFVG SKRLRAGDSV LFIRDEKSQL 240 RVGVRRANRQ QTTLPSSVLS ADSMHIGVLA AAAHAAANRS PFTIFYNPRA CPSDFVIPLA 300 KYRKSVYGTQ LSAGMRFGMM FETEESGKRR YMGTIVGISD VDPLRWPGSK WRNIQAEWDE 360 PGCGDKQNRV SVWEIETPES LFIFPSLTSS LKRPLQSGYL ENEWGTLIRR PFIKVPENGG 420 TVEISSSMQN LYQENMKMFY KPQVINNSGA FLPIMQQDYS PTRVPFQDMK STQALENPKV 480 HLSSTESIPN NTFNMHSLLK NDQPEKLQPL AKTDNSKLES EVLPDHIFDF PSFESCSNIE 540 KVNTANQLTY HNQNQSQNPL LAHTSPWTMQ QPQLESSMSH PQLIDMVQPD SSIVNSMLPQ 600 LDVDEWMMYS SCQPQVVNPS IPSMNQEVWD QYVKNFNLRD LSAESNNQSE ICVNVDASNS 660 VSTTVVDPST SNTIFDDFCS MKDKDFQHPQ DCMVGNLSSS QDGQSQITSA SLAESHAFSL 720 RDNSGGTSSS HVDFDESSFL QNNNSWKQVA APIRTYTKVQ KAGSVGRSID VTTFKNYEEL 780 IRAIECMFGL DGLLNDTKGS GWKLVYVDYE SDVLLVGDDP WEEFVGCVRC IRILSPSEVK 840 EMSEEGMKLL NSGALQGINV DEKSPLRVGV RRANRQQTTL PSSVLSADSM HIGVLAAAAH 900 AAANRIPFTI FYNPRACLSD FVIPLAKSVY GTQLSVGMRF GCLRPRNRAE WDEPGCGDKQ 960 NRLQPLAKTD NSKLESEVLP DHMFDFPSFE SCSNIEKLES SMSHPQLIDM VQPDSSIANS 1020 MLPQLDVDEW MMYSSCQPQV VNPSIPSMNQ EMWDQYVKNF NLRDLSAESN NQSEICVNVD 1080 ATNSVSTTMV DPSTTNTIFD EFCSMKDKDF QHPQDCAVGN LSSSQDGQSQ ITSASLAESH 1140 AFSLRDNSGG TSSSHVDFDE SIFLQNNNSW KQVAAPIRTY TKLEKAGSVG RSIDVTTFKN 1200 YEELIRAIEY MFGLDGLLND TKDDPWKEFV GCVRRIRILS PSEVKEMSEE GMKLLNSGAL 1260 QGINV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 0.0 | 19 | 965 | 32 | 392 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.25092 | 0.0 | root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: In early embryo and during organ development. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the whole plant with a lower expression in leaves. Detected in embryo axis, provascular tissues, procambium and some differentiated vascular regions of mature organs. {ECO:0000269|PubMed:10476078}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Mediates embryo axis formation and vascular tissues differentiation. Functionally redundant with ARF7. May be necessary to counteract AMP1 activity. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:14973283, ECO:0000269|PubMed:17553903}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00153 | DAP | Transfer from AT1G19850 | Download |
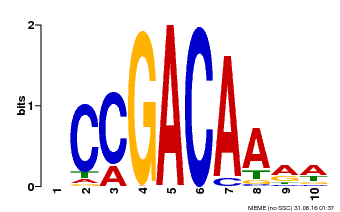
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr1g024025.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024631194.1 | 0.0 | auxin response factor 5 | ||||
| Refseq | XP_024631195.1 | 0.0 | auxin response factor 5 | ||||
| Swissprot | P93024 | 0.0 | ARFE_ARATH; Auxin response factor 5 | ||||
| TrEMBL | A0A072VFI0 | 0.0 | A0A072VFI0_MEDTR; Auxin response factor | ||||
| STRING | XP_004499235.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8991 | 32 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19850.1 | 0.0 | ARF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr1g024025.1 |
| Entrez Gene | 25482193 |



