 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr1g050502.2 | ||||||||
| Common Name | MTR_1g050502 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 213aa MW: 23900.9 Da PI: 4.4606 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 38.5 | 2.6e-12 | 105 | 149 | 3 | 47 |
XXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 3 elkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkk 47
+k+++r ++NR AA +sR+RKk ++++Le+k+ +e+e ++L
Medtr1g050502.2 105 VSKKQIRQMRNRDAAVKSRERKKVYVKNLETKSRYFEGECRRLEH 149
579************************************987754 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 3.0E-12 | 98 | 165 | No hit | No description |
| SMART | SM00338 | 7.6E-5 | 103 | 167 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 8.542 | 105 | 148 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 8.7E-10 | 106 | 149 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.63E-10 | 107 | 163 | No hit | No description |
| CDD | cd14704 | 3.21E-16 | 108 | 159 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 111 | 125 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0030968 | Biological Process | endoplasmic reticulum unfolded protein response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 213 aa Download sequence Send to blast |
MDTFEPQIDF ESLFAEYEVA DFLQEDDINP SVSVADTPSP LSEIENLLMS DAEGIASSSS 60 DSDYHKLLQD ILVDPIPLSD DQGFVLSDEA RVDPVTPQEV PPEPVSKKQI RQMRNRDAAV 120 KSRERKKVYV KNLETKSRYF EGECRRLEHL LQCCYAENHA LRLCLQSRGG FGAPMTMQES 180 AVLLLGKNPC CWVPCYGSWA SCANSVYPFH CR* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.3000 | 0.0 | leaf| root | ||||
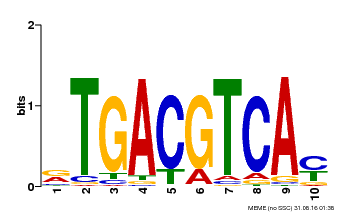
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00019 | PBM | Transfer from AT1G42990 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr1g050502.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT138826 | 2e-98 | BT138826.1 Lotus japonicus clone JCVI-FLLj-17J20 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013467378.1 | 1e-132 | bZIP transcription factor 60 | ||||
| TrEMBL | A0A072VHD6 | 1e-156 | A0A072VHD6_MEDTR; BZIP transcription factor | ||||
| STRING | XP_004514723.1 | 6e-76 | (Cicer arietinum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G42990.1 | 2e-15 | basic region/leucine zipper motif 60 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr1g050502.2 |
| Entrez Gene | 25483259 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



