 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr2g101190.1 | ||||||||
| Common Name | MTR_2g101190 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 837aa MW: 92138.5 Da PI: 6.5764 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57.2 | 2.8e-18 | 14 | 72 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++ +++ps +r++L +++ +++ +q+kvWFqNrR +ek+
Medtr2g101190.1 14 KYVRYTPEQVEALERLYHDCPKPSSIRRQQLIRECpilsHIEPKQIKVWFQNRRCREKQ 72
5679*****************************************************97 PP
| |||||||
| 2 | START | 175.8 | 2.6e-55 | 158 | 366 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EEEEE CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..galql 93
+aee+++e+++ka+ ++ Wv+++ +++g+e++ +++ s++++g a+ra+g+v +++ v+e+l+d++ W ++++++++l+v+ ++ g+++l
Medtr2g101190.1 158 IAEETLAEFLSKATGTAVEWVQMPGMKPGPESIGIIAISHGCHGVAARACGLVGLEPT-RVAEILKDRPSWYRDCRAVDILNVLPTAngGTIEL 250
7899******************************************************.8888888888****************9999***** PP
EEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHHHHHH CS
START 94 mvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlphwllrs 183
+++l+a+++l+p Rd + +Ry+ l++g++vi+++S+ ++q+ p+ +++vRae+lpSg+li+p+++g+s +++v+h+dl+ ++++++lr+
Medtr2g101190.1 251 LYMQLYAPTTLAPaRDLWLLRYTSVLEDGSLVICERSLKNTQNGPSmppVQHFVRAEMLPSGYLIRPCEGGGSIIHIVDHMDLEPWSVPEVLRP 344
********************************************9988899******************************************* PP
HHHHHHHHHHHHHHHHTXXXXX CS
START 184 lvksglaegaktwvatlqrqce 205
l++s+++ ++kt++ +l+++++
Medtr2g101190.1 345 LYESSTMLAQKTTMVALRHLRQ 366
**************99998875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 15.111 | 9 | 73 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.3E-15 | 11 | 77 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 9.41E-17 | 14 | 77 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 3.87E-16 | 14 | 74 | No hit | No description |
| Pfam | PF00046 | 8.0E-16 | 15 | 72 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 9.5E-18 | 16 | 72 | IPR009057 | Homeodomain-like |
| CDD | cd14686 | 1.71E-6 | 66 | 105 | No hit | No description |
| PROSITE profile | PS50848 | 24.677 | 148 | 349 | IPR002913 | START domain |
| CDD | cd08875 | 1.57E-78 | 152 | 367 | No hit | No description |
| Gene3D | G3DSA:3.30.530.20 | 2.3E-22 | 157 | 363 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 1.19E-36 | 157 | 366 | No hit | No description |
| SMART | SM00234 | 1.1E-41 | 157 | 367 | IPR002913 | START domain |
| Pfam | PF01852 | 1.1E-52 | 158 | 366 | IPR002913 | START domain |
| Pfam | PF08670 | 2.0E-49 | 693 | 835 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 837 aa Download sequence Send to blast |
MSCKDGKGVM DNGKYVRYTP EQVEALERLY HDCPKPSSIR RQQLIRECPI LSHIEPKQIK 60 VWFQNRRCRE KQRKEASRLQ AVNRKLSAMN KLLMEENDRL QKQVSHLVYE NGYFRQHTQN 120 TNLATKDTSC DSAVTSGQRS LTAQHPPRDA SPAGLLSIAE ETLAEFLSKA TGTAVEWVQM 180 PGMKPGPESI GIIAISHGCH GVAARACGLV GLEPTRVAEI LKDRPSWYRD CRAVDILNVL 240 PTANGGTIEL LYMQLYAPTT LAPARDLWLL RYTSVLEDGS LVICERSLKN TQNGPSMPPV 300 QHFVRAEMLP SGYLIRPCEG GGSIIHIVDH MDLEPWSVPE VLRPLYESST MLAQKTTMVA 360 LRHLRQISHE VSQSNVTGWG RRPAALRALG QRLSRGFNEA LNGFTDEGWS MIGNDGVDDV 420 TILVNSSPDK LMGLNLSFAN GFPSVSNAVL CAKASMLLQN VPPAILLRFL REHRSEWADN 480 NMDAYSAAAI KAGPCSFSGS RVGNYGGQVI LPLAHTIEHE EFLEVIKLEG IAHSPEDAIM 540 PREVFLLQLC SGMDENAVGT CAELIFAPID ASFADDAPLL PSGFRIIPLD SGKEVANNPN 600 RTLDLTSALD IGPAGNKASN DYSGNSGCMR SVMTIAFEFA FESHMQDHVA SMARQYVRSI 660 ISSVQRVALA LSPSNLNSQG GLRTPLGTPE AQTLARWISN SYRCFLGAEL LKSNNEGSES 720 LLKSLWHHTD AILCCTLKAL PVFTFANQAG LDMLETTLVA LQDIALEKIF DDHGRKILCS 780 EFPLIIQQGF ACLQGGLCLS SMGRPISYER VVAWKVLNEE ENAHCICFMF VNWSFV* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.8412 | 0.0 | leaf| root| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Highly expressed the developing vascular elements and the adaxial portion of cotyledons. Expressed in developing ovules, stamens and carpels. Expressed in procambium and shoot meristem. {ECO:0000269|PubMed:14701930, ECO:0000269|PubMed:15705957}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulates procambial and vascular tissue formation or maintenance, and vascular development in inflorescence stems. {ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:15705957, ECO:0000269|PubMed:16617092}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
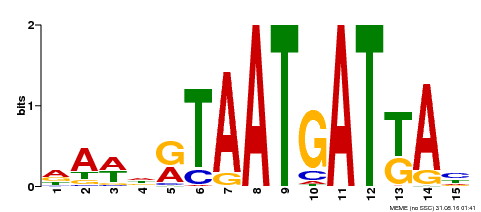
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr2g101190.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:15773855, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:16617092, ECO:0000269|PubMed:17237362}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003597690.1 | 0.0 | homeobox-leucine zipper protein ATHB-15 | ||||
| Swissprot | Q9ZU11 | 0.0 | ATB15_ARATH; Homeobox-leucine zipper protein ATHB-15 | ||||
| TrEMBL | G7II12 | 0.0 | G7II12_MEDTR; Class III HD-Zip protein CNA1 | ||||
| STRING | AES67941 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1946 | 33 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.2 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr2g101190.1 |
| Entrez Gene | 11432376 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



