 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr3g065980.1 | ||||||||
| Common Name | MTR_3g065980 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | GRAS | ||||||||
| Protein Properties | Length: 548aa MW: 60002.6 Da PI: 4.7669 | ||||||||
| Description | GRAS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GRAS | 459.4 | 2.2e-140 | 169 | 540 | 1 | 374 |
GRAS 1 lvelLlecAeavssgdlelaqalLarlselaspdgdpmqRlaayfteALaarlarsvselykalppsetseknsseelaalklfsevsPilkfs 94
lv++L++cAea+++++l+la+al++++s las + +m+++a+yf++ALa+r+++ +p+et +++ se l++ f+e sP+lkf+
Medtr3g065980.1 169 LVHTLMACAEAIQQKNLKLAEALVKHISLLASLQTGAMRKVASYFAQALARRIYG---------NPEETIDSSFSEILHM--HFYESSPYLKFA 251
689****************************************************.........7777776566666666..5*********** PP
GRAS 95 hltaNqaIleavegeervHiiDfdisqGlQWpaLlqaLasRpegppslRiTgvgspesgskeeleetgerLakfAeelgvpfefnvlvakrled 188
h+taNqaIlea++g+ rvH+iDf+++qG+QWpaL+qaLa Rp+gpp++R+Tg+g+p+++++++l+++g++La++A+++gv+fef+ +v+++++d
Medtr3g065980.1 252 HFTANQAILEAFAGAGRVHVIDFGLKQGMQWPALMQALALRPGGPPTFRLTGIGPPQADNTDALQQVGWKLAQLAQTIGVQFEFRGFVCNSIAD 345
********************************************************************************************** PP
GRAS 189 leleeLrvkpgEalaVnlvlqlhrlldesvsleserdevLklvkslsPkvvvvveqeadhnsesFlerflealeyysalfdsleak........ 274
l++++L+++pgEa+aVn+v++lh++l++++s+e+ vL++vk+++Pk+v++veqea+hn++ F++rf+eal+yys+lfdsle +
Medtr3g065980.1 346 LDPNMLEIRPGEAVAVNSVFELHTMLARPGSVEK----VLNTVKKINPKIVTIVEQEANHNGPVFVDRFTEALHYYSSLFDSLEGSnsssnnsn 435
**********************************....*********************************************77767777766 PP
GRAS 275 ...lpres....eerikvErellgreivnvvacegaerrerhetlekWrerleeaGFkpvplsekaakqaklllrkvk.sdgyrveeesgslvl 360
+++ +++ +lg++i+nvva eg +r+erhetl +Wr+r+++aGF+pv+l+++a kqa++ll+ ++ +dgyrvee++g+l+l
Medtr3g065980.1 436 snsT---GlgspSQDLLMSEIYLGKQICNVVAYEGVDRVERHETLTQWRSRMGSAGFEPVHLGSNAFKQASTLLALFAgGDGYRVEENNGCLML 526
5552...0444788999999************************************************************************** PP
GRAS 361 gWkdrpLvsvSaWr 374
gW++r+L+++SaW+
Medtr3g065980.1 527 GWHTRSLIATSAWK 540
*************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01129 | 1.0E-34 | 14 | 87 | No hit | No description |
| Pfam | PF12041 | 4.2E-36 | 14 | 80 | IPR021914 | Transcriptional factor DELLA, N-terminal |
| PROSITE profile | PS50985 | 67.892 | 143 | 519 | IPR005202 | Transcription factor GRAS |
| Pfam | PF03514 | 7.7E-138 | 169 | 540 | IPR005202 | Transcription factor GRAS |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042176 | Biological Process | regulation of protein catabolic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:2000033 | Biological Process | regulation of seed dormancy process | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 548 aa Download sequence Send to blast |
MWREEKETNG GGMDELLAAL GYKVRSSDMA DVAQKLEQLE MVMGSAQEEG INHLSSDTVH 60 YDPTDLYSWV QTMLTELNPD SSQINDPLAS LGSSSEILNN TFNDDSEYDL SAIPGMAAYP 120 PQEENTAAKR MKTWSEPESE PAVVMSPPPA VENTRPVVLV DTQETGVRLV HTLMACAEAI 180 QQKNLKLAEA LVKHISLLAS LQTGAMRKVA SYFAQALARR IYGNPEETID SSFSEILHMH 240 FYESSPYLKF AHFTANQAIL EAFAGAGRVH VIDFGLKQGM QWPALMQALA LRPGGPPTFR 300 LTGIGPPQAD NTDALQQVGW KLAQLAQTIG VQFEFRGFVC NSIADLDPNM LEIRPGEAVA 360 VNSVFELHTM LARPGSVEKV LNTVKKINPK IVTIVEQEAN HNGPVFVDRF TEALHYYSSL 420 FDSLEGSNSS SNNSNSNSTG LGSPSQDLLM SEIYLGKQIC NVVAYEGVDR VERHETLTQW 480 RSRMGSAGFE PVHLGSNAFK QASTLLALFA GGDGYRVEEN NGCLMLGWHT RSLIATSAWK 540 LPQNESK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_A | 3e-67 | 164 | 539 | 13 | 377 | Protein SCARECROW |
| 5b3h_D | 3e-67 | 164 | 539 | 13 | 377 | Protein SCARECROW |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.4298 | 0.0 | glandular trichome| leaf| pod| root| seed| stem | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional regulator that acts as a repressor of the gibberellin (GA) signaling pathway. Probably acts by participating in large multiprotein complexes that represses transcription of GA-inducible genes. Upon GA application, it is degraded by the proteasome, allowing the GA signaling pathway (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00611 | ChIP-seq | Transfer from AT2G01570 | Download |
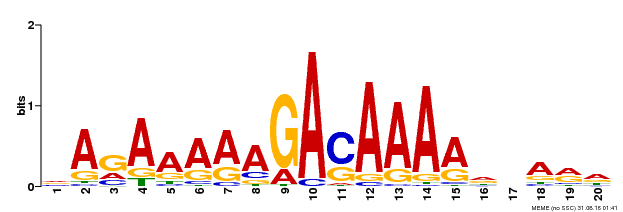
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr3g065980.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013460591.2 | 0.0 | DELLA protein GAI | ||||
| Swissprot | Q84TQ7 | 0.0 | GAI_GOSHI; DELLA protein GAI | ||||
| TrEMBL | A0A396IUP1 | 0.0 | A0A396IUP1_MEDTR; Putative transcription factor GRAS family | ||||
| STRING | XP_004503135.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF803 | 34 | 121 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01570.1 | 0.0 | GRAS family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr3g065980.1 |
| Entrez Gene | 25489240 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



