 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr3g077750.1 | ||||||||
| Common Name | MTR_3g077750 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 337aa MW: 37269.5 Da PI: 7.1674 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 126.4 | 8.7e-40 | 59 | 117 | 4 | 62 |
zf-Dof 4 kalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
+lkcprCdst+tkfCyynnyslsqPryfCk+CrryWtkGG+lrn+PvGgg+rknkk+s
Medtr3g077750.1 59 LSLKCPRCDSTHTKFCYYNNYSLSQPRYFCKTCRRYWTKGGTLRNIPVGGGCRKNKKVS 117
5689****************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 2.0E-28 | 51 | 111 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 1.8E-33 | 60 | 115 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.189 | 61 | 115 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 63 | 99 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010067 | Biological Process | procambium histogenesis | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 337 aa Download sequence Send to blast |
MGLSSLHVCN MESSADHWLQ GTIHDESGMD SSSPMSGDML TCSRPSSMIE RRLRPPHDLS 60 LKCPRCDSTH TKFCYYNNYS LSQPRYFCKT CRRYWTKGGT LRNIPVGGGC RKNKKVSSKK 120 SNDHLANTNI QNQPHHVPSY HQNPKDLQLS FPDVQFSHLS NLFGANGALG NPNFMENKYS 180 NVGMLENPRP IDFMMENKLE GIIGSSSRNF DNYFGNSDHM NMNMGVGIGG DMMNGQNGLP 240 QNFHHAFGGM SFEGGNNNNG AYLMDSCQRL MLPYDANDED HNASIDVKPN PKLLSLEWQQ 300 DQGCSDAGKG QSFGYGSWSG MMNGYGSSTT NPLING* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.17476 | 0.0 | root| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in the vascular tissues and pericycle of primary roots. Detected in the vasculature of the cotyledons, rosette leaves and cauline leaves. In flowers, localized in vasculature of petals, stigma, and stamen filaments, and, to a lower extent, in anthers and carpels. In inflorescence stems, accumulates in the vasculature, particularly in cambium, phloem, and interfascicular parenchyma cells. {ECO:0000269|PubMed:19915089}. | |||||
| Uniprot | TISSUE SPECIFICITY: The PEAR proteins (e.g. DOF2.4, DOF5.1, DOF3.2, DOF1.1, DOF5.6 and DOF5.3) form a short-range concentration gradient that peaks at protophloem sieve elements (PSE) (PubMed:30626969). Preferentially expressed in the vasculature of all organs, including seedlings, roots, stems, buds, leaves, flowers and siliques, and particularly in the cambium, phloem and interfascicular parenchyma cells of inflorescence stems (PubMed:19915089). {ECO:0000269|PubMed:19915089, ECO:0000269|PubMed:30626969}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence (By similarity). Promotes expression (PubMed:19915089). The PEAR proteins (e.g. DOF2.4, DOF5.1, DOF3.2, DOF1.1, DOF5.6 and DOF5.3) activate gene expression that promotes radial growth of protophloem sieve elements (PubMed:30626969). Involved in the regulation of interfascicular cambium formation and vascular tissue development, particularly at a very early stage during inflorescence stem development; promotes both cambium activity and phloem specification, but prevents xylem specification (PubMed:19915089). {ECO:0000250|UniProtKB:Q9M2U1, ECO:0000269|PubMed:19915089, ECO:0000269|PubMed:30626969}. | |||||
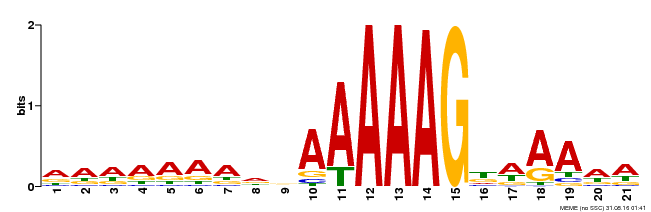
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00580 | DAP | Transfer from AT5G62940 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr3g077750.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CT954236 | 0.0 | CT954236.4 M.truncatula DNA sequence from clone MTH2-3H12 on chromosome 3, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003601260.2 | 0.0 | dof zinc finger protein DOF5.6 | ||||
| Swissprot | Q9FM03 | 6e-55 | DOF56_ARATH; Dof zinc finger protein DOF5.6 | ||||
| TrEMBL | G7J5F5 | 0.0 | G7J5F5_MEDTR; Dof domain zinc finger protein | ||||
| STRING | AES71511 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5092 | 34 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62940.1 | 1e-52 | Dof family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr3g077750.1 |
| Entrez Gene | 11430187 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



