 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr3g084720.1 | ||||||||
| Common Name | MTR_3g084720 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 230aa MW: 26066.4 Da PI: 9.8839 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 15.9 | 3.6e-05 | 134 | 157 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirt.H 23
y+C++C+k F ++ + +H ++ H
Medtr3g084720.1 134 YPCSKCNKIFETSQKFANHVSSsH 157
99*****************99988 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50157 | 9.349 | 134 | 162 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 136 | 157 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 230 aa Download sequence Send to blast |
MNNNRLFQWM HGYNTTLKIY THSSFDYDSS SSVGTNNFGS HAPGILKAME ILETQFGHLH 60 TSTSSSSSSI NINMNDDIII NNNKNNNMMM TMNTYVAASS HLDLHNDMRK KKKRNENIND 120 GRIHSLPHKK YGPYPCSKCN KIFETSQKFA NHVSSSHCKF ESEEDRKKRY ISRIRKRPRL 180 QIQKLNDGTT TFVPVIACGD KSHAYVNDDG HNMIALTPLP NGIKVKSEP* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 108 | 113 | RKKKKR |
| 2 | 172 | 178 | RIRKRPR |
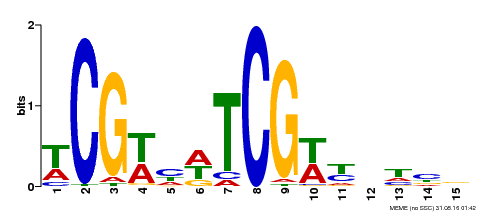
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00523 | DAP | Transfer from AT5G22990 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr3g084720.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC147484 | 0.0 | AC147484.21 Medicago truncatula clone mth2-52b4, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024633575.1 | 3e-96 | probable serine/threonine-protein kinase DDB_G0271402 | ||||
| TrEMBL | G7JBB9 | 1e-170 | G7JBB9_MEDTR; Uncharacterized protein | ||||
| STRING | AES71974 | 1e-123 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF16742 | 7 | 7 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G22990.1 | 3e-15 | C2H2-like zinc finger protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr3g084720.1 |
| Entrez Gene | 11438517 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



