 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr3g100050.1 | ||||||||
| Common Name | MTR_3g100050 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 310aa MW: 34206.5 Da PI: 5.8294 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 50.8 | 2.2e-16 | 200 | 236 | 1 | 35 |
GATA 1 CsnCgttk..TplWRrgpdgnktLCnaCGlyyrkkgl 35
C++Cg ++ Tp++Rrgp+g++tLCnaCGl+++ +g+
Medtr3g100050.1 200 CTHCGISSksTPMMRRGPSGPRTLCNACGLFWANRGT 236
*****99999***********************9997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00979 | 2.3E-10 | 65 | 100 | IPR010399 | Tify domain |
| PROSITE profile | PS51320 | 14.911 | 65 | 100 | IPR010399 | Tify domain |
| Pfam | PF06200 | 1.5E-12 | 69 | 99 | IPR010399 | Tify domain |
| Pfam | PF06203 | 2.0E-15 | 132 | 174 | IPR010402 | CCT domain |
| PROSITE profile | PS51017 | 12.878 | 132 | 174 | IPR010402 | CCT domain |
| SMART | SM00401 | 8.2E-15 | 194 | 247 | IPR000679 | Zinc finger, GATA-type |
| PROSITE profile | PS50114 | 10.238 | 194 | 250 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 2.33E-12 | 197 | 248 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 6.7E-16 | 198 | 241 | IPR013088 | Zinc finger, NHR/GATA-type |
| CDD | cd00202 | 9.63E-15 | 199 | 241 | No hit | No description |
| Pfam | PF00320 | 5.5E-14 | 200 | 236 | IPR000679 | Zinc finger, GATA-type |
| PROSITE pattern | PS00344 | 0 | 200 | 227 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 310 aa Download sequence Send to blast |
MYNSMNMSDQ IKNNEDPDNN NNHIHFDSPT LEDASGEGEG QINDVSLENV YVSGDGNHPD 60 MSVQQFDDSS QLTLSFRGQV YVFDSVTPEK VQSVLLLLGG CELNPGSQCL DTSPLNQRSG 120 AEFPTRCSQP QRAASLIRFR QKRKERNFDK KVRYEVRQEV ALRMQRSKGQ FTSAKKQDGG 180 NSWGSDPESG QDVVQSETSC THCGISSKST PMMRRGPSGP RTLCNACGLF WANRGTLRDL 240 STARRNHEQH TLGSPEQGMR DLSNPKRNHQ PHPLPQPEQV GEGNEDLNCG TAPAHNDSVD 300 DKTAVVSDQ* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.50 | 0.0 | pod| root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Predominantly expressed in shoot apices, inflorescences and roots. {ECO:0000269|PubMed:10945256, ECO:0000269|PubMed:14966217}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. {ECO:0000250, ECO:0000269|PubMed:14966217}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00451 | DAP | Transfer from AT4G24470 | Download |
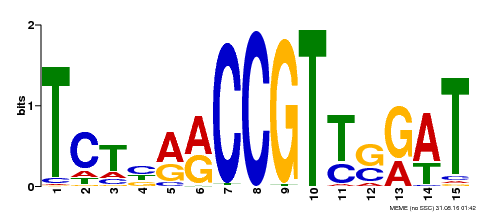
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr3g100050.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT138662 | 0.0 | BT138662.1 Medicago truncatula clone JCVI-FLMt-15G4 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003602888.1 | 0.0 | GATA transcription factor 25 | ||||
| Swissprot | Q9LRH6 | 3e-75 | GAT25_ARATH; GATA transcription factor 25 | ||||
| TrEMBL | G7J4K3 | 0.0 | G7J4K3_MEDTR; GATA transcription factor | ||||
| STRING | AES73139 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2836 | 33 | 68 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G24470.3 | 2e-77 | GATA-type zinc finger protein with TIFY domain | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr3g100050.1 |
| Entrez Gene | 11420876 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



