 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr3g101780.2 | ||||||||
| Common Name | MTR_3g101780 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 432aa MW: 47142 Da PI: 9.8401 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 47.3 | 4.6e-15 | 353 | 404 | 5 | 56 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkev 56
+r+rr++kNRe+A rsR+RK+a++ eLe +v++L++eN++L+k+ ee+ ++
Medtr3g101780.2 353 RRQRRMIKNRESAARSRARKQAYTMELEAEVAKLKEENEELQKKQEEIMELQ 404
69****************************************9999988765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF81995 | 1.36E-5 | 198 | 263 | No hit | No description |
| SMART | SM00338 | 5.7E-14 | 349 | 414 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.519 | 351 | 402 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.38E-10 | 353 | 402 | No hit | No description |
| CDD | cd14707 | 3.21E-28 | 353 | 407 | No hit | No description |
| Pfam | PF00170 | 3.9E-13 | 353 | 405 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 1.5E-14 | 353 | 403 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 356 | 371 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0010255 | Biological Process | glucose mediated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 432 aa Download sequence Send to blast |
MNFKGFGNDP GASGNGGGRI AAGNFPLTRQ PSVYSLTVDE FMNSMGGSGK DFGSMNMDEL 60 LKNIWSAEEV QTMGGEEAIS NHLQRQGSLT LPRTLSQKTV DEVWKDISKD YGGPNLAAPM 120 TQRQPTLGEM TLEEFLVRAG VVREDAKPND GVFLDLGNVG NNGNLGLAFQ AQQMNKVAGF 180 MGNGNRINGN DDPLVGLQSP TNLPLNVNGI RSTNQQQQMQ NSQSQAQQQH QNQQLQQLQQ 240 QQQQQQIFPK QPGLNYATQM PLSNNQGMRG GIVGLSPDHG MNGNLVQGGG IGMVGLAPGA 300 VQIGAVSPAN QISSDKMGKS NGDTSSVSPV PYVFNGGMRG RKGNGAVEKV IERRQRRMIK 360 NRESAARSRA RKQAYTMELE AEVAKLKEEN EELQKKQEEI MELQKNQVKE MMNLQREVKR 420 KCLRRTQTGP W* |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, leaves, flowers and siliques but not in seeds. {ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in ABA and stress responses and acts as a positive component of glucose signal transduction. Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. {ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00186 | DAP | Transfer from AT1G45249 | Download |
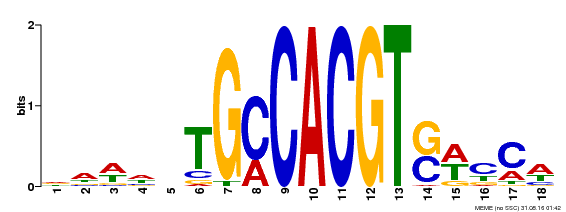
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr3g101780.2 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA), cold and glucose. {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CU012043 | 0.0 | CU012043.5 M.truncatula DNA sequence from clone MTH2-63O12 on chromosome 3, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003603049.1 | 0.0 | bZIP transcription factor 46 | ||||
| Refseq | XP_024635171.1 | 0.0 | bZIP transcription factor 46 | ||||
| Refseq | XP_024635172.1 | 0.0 | bZIP transcription factor 46 | ||||
| Swissprot | Q9M7Q4 | 1e-123 | AI5L5_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| TrEMBL | G7J6E3 | 0.0 | G7J6E3_MEDTR; ABA response element-binding factor | ||||
| STRING | AES73300 | 0.0 | (Medicago truncatula) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G45249.1 | 1e-104 | abscisic acid responsive elements-binding factor 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr3g101780.2 |
| Entrez Gene | 11435200 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



