 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr3g116070.1 | ||||||||
| Common Name | MTR_3g116070 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 354aa MW: 40385.7 Da PI: 7.142 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 167.1 | 5.9e-52 | 23 | 151 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevls 94
lppGfrFhPtdeel+++yL+ kv +++ + ++evd++++ePw+Lp+++k +e+ewy Fs rd+ky+tg r+nrat++gyWkatgkdkev+s
Medtr3g116070.1 23 LPPGFRFHPTDEELITFYLASKVFKNTYFNNVKFAEVDLNRCEPWELPEMAKMGEREWYLFSLRDRKYPTGLRTNRATSAGYWKATGKDKEVYS 116
79************************9666455***************9889999**************************************9 PP
NAM 95 k.kgelvglkktLvfykgrapkgektdWvmheyrl 128
+++l g+kktLvfykgrap+gekt+Wvmheyrl
Medtr3g116070.1 117 CsSRSLLGMKKTLVFYKGRAPRGEKTKWVMHEYRL 151
756777***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 5.89E-56 | 18 | 158 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 53.193 | 23 | 167 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.8E-26 | 24 | 151 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010199 | Biological Process | organ boundary specification between lateral organs and the meristem | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 354 aa Download sequence Send to blast |
MMLAMEEVLC ELSDHEKKNE QGLPPGFRFH PTDEELITFY LASKVFKNTY FNNVKFAEVD 60 LNRCEPWELP EMAKMGEREW YLFSLRDRKY PTGLRTNRAT SAGYWKATGK DKEVYSCSSR 120 SLLGMKKTLV FYKGRAPRGE KTKWVMHEYR LHSHHFSPSN TCKEEKKRNS LLQIQGHLGG 180 NPNLSPQKSC LPPTPLLLAT SFTQLDQNDQ LLSHSHNIFP LPTFQPSFSM INHTTRNNNN 240 SPSSELLFKS QQNYTMKQTI PKTEATFYDQ QKSIDDALNI RWIIDNNNNY ENSLFPVEME 300 MEMEMDGAAH DLIAFSGAAT NAEFRDISII NSRGGGGVVG PMGIDSWPHA QLV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 1e-44 | 14 | 151 | 6 | 140 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00241 | DAP | Transfer from AT1G76420 | Download |
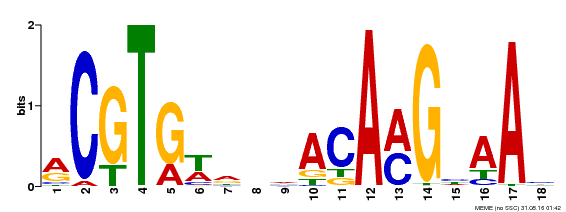
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr3g116070.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC131026 | 0.0 | AC131026.11 Medicago truncatula clone mth2-6e18, complete sequence. | |||
| GenBank | AC144482 | 0.0 | AC144482.13 Medicago truncatula clone mth2-10p1, complete sequence. | |||
| GenBank | CU571057 | 0.0 | CU571057.2 M.truncatula DNA sequence from clone MTH2-52O4 on chromosome 3, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024633445.1 | 0.0 | protein CUP-SHAPED COTYLEDON 3 | ||||
| TrEMBL | G7J6G2 | 0.0 | G7J6G2_MEDTR; CUP-shaped cotyledon-like protein | ||||
| STRING | AES74121 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7888 | 33 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76420.1 | 2e-87 | NAC family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr3g116070.1 |
| Entrez Gene | 11422778 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



